This article dives into a comparative benchmark of two advanced language models, both tested on a wide array of clinical and biomedical language understanding tasks.
On one side stands Claude 3.7 Sonnet from Anthropic, which represents the frontier model most widely used in their suite. On the other is John Snow Labs’ domain-specific suite of healthcare LLMs, available in several sizes such as 7B, 10B, 14B, 24B, and larger.
What matters when evaluating LLMs for medical use
Over time, the research community has reached agreement that medical AI systems must be judged on several criteria: factual correctness, relevance in clinical practice, completeness, safety, clarity, and resistance to hallucination.
The evaluation the results of which we present here involved over 200 newly developed prompts across four clinical task types: summarizing medical notes, extracting structured data from clinical text, answering research-level questions, and general open-ended medical queries. All prompts were newly written to ensure that none of the tested models had previously encountered the material.
Evaluating medical LLMs demands expert-based benchmarks focused on factual accuracy and clinical relevance in a randomized and blinded experimental design.
Physician reviewers assessed each output in a randomized and blinded setup, comparing anonymized responses from both models. They could select a preferred response or declare a tie. The process was designed with inter-rater agreement analysis and included re-assessments two weeks later to check for consistency in ratings.
The core metrics used in this evaluation were factuality (whether the output was medically correct), clinical relevance (whether a physician would find the response informative), and conciseness (whether the message was delivered succinctly). This report centers on the first two, since conciseness is more vulnerable to prompt tweaks.
Across the board, John Snow Labs’ LLM surpasses Claude 3.7 Sonnet
When comparing outputs for all evaluated tasks, physician raters showed no strong preference in 67% of cases when judging factuality. But when they did express a choice, they leaned toward John Snow Labs’ Medical LLM 21% of the time, versus 12% for Claude 3.7 Sonnet. Regarding clinical relevancy, the distribution of neutral ratings was 54%, but when raters made a call, the John Snow Labs model was selected in 24% of cases compared to GPT-4.5’s 22%.
John Snow Labs’ Medical LLM was preferred nearly twice as often as Claude 3.7 Sonnet for factual accuracy, at only 20% of the cost.
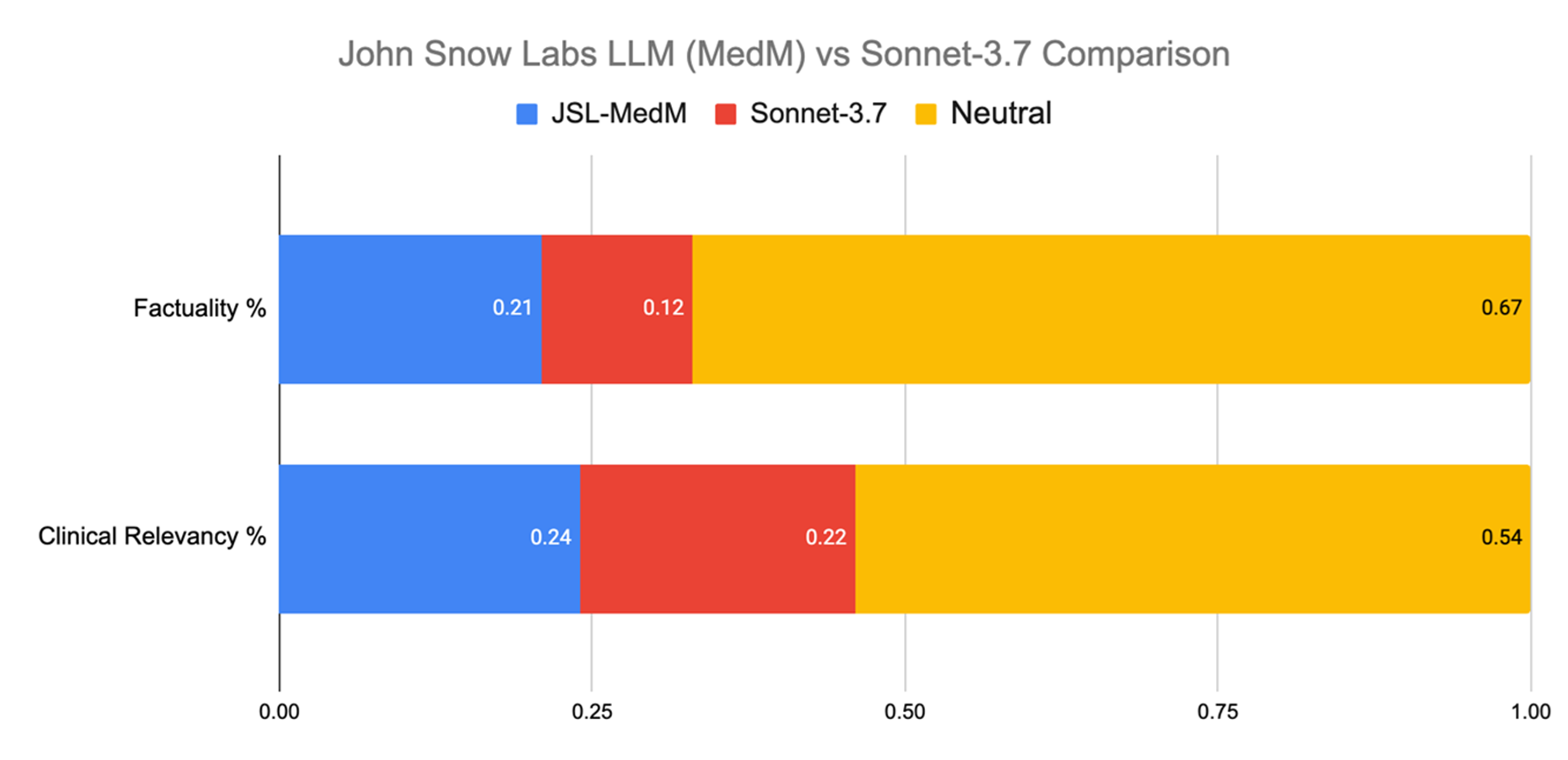
Evaluation across all tasks.
Most notably, this performance came at just 20% of Claudes 3.7 Sonnet’s operating cost, taking into account infrastructure, deployment, and licensing expenses.
Clinical Data Extraction
Extracting information from clinical texts is essential for many workflows, such as identifying interventions in imaging reports or summarizing patient background from intake forms. These capabilities support tasks ranging from pharmacovigilance to public health.
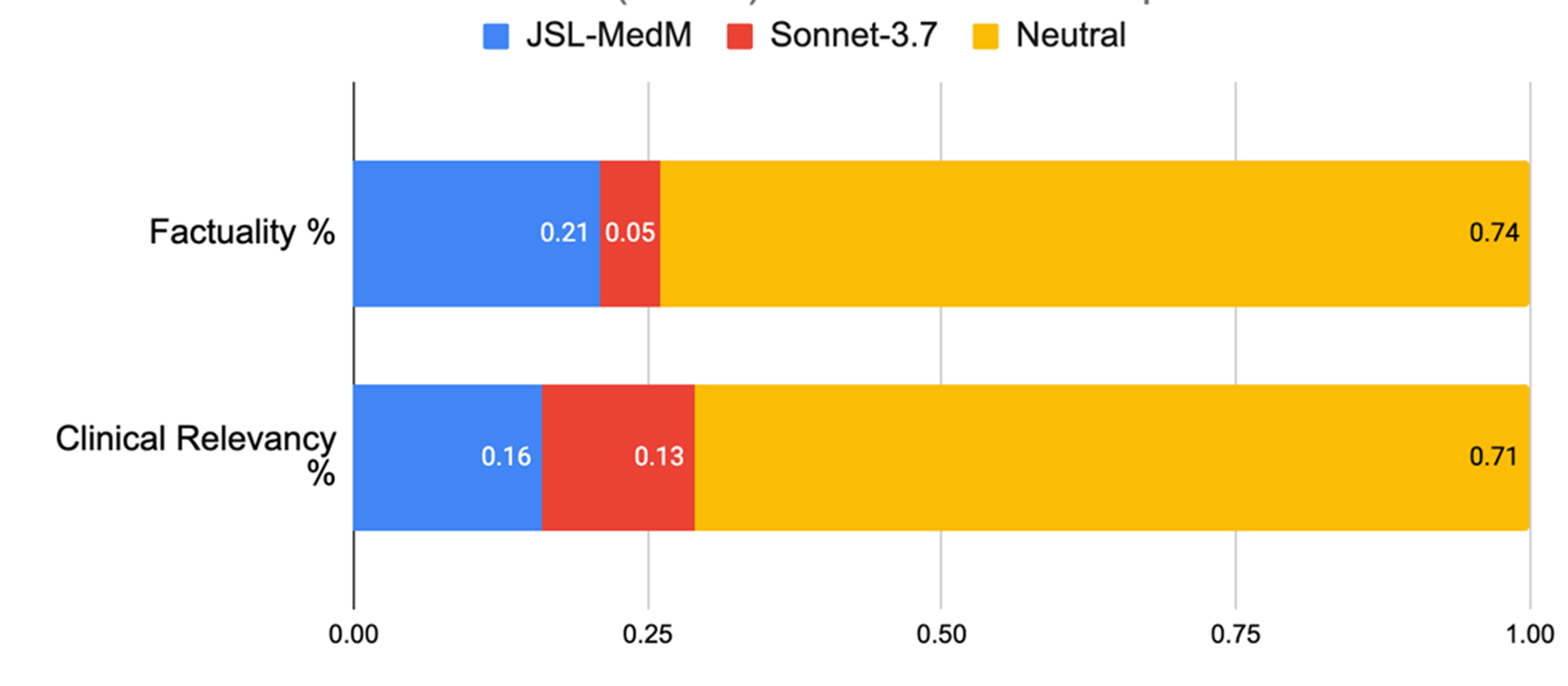
Evaluation on Clinical Information Extraction.
In this area, both models were judged equally accurate 74% of the time. But when raters expressed a preference, John Snow Labs’ model was selected four times more frequently than Claude 3.7 Sonnet (21% vs 5%). For clinical relevance, it again pulled ahead, favored 16% of the time compared to 13% for Claude 3.7 Sonnet.
In clinical data extraction, John Snow Labs’ model was chosen four times more often than Claude 3.7 Sonnet when experts expressed a preference.
Biomedical Q&A
This task included questions like “What biomarkers are commonly negative in APL?” or “What is sNFL used for?” In terms of producing medically relevant responses, the John Snow Labs model was preferred over Claude 3.7. Sonnet in 25% of cases versus 22%. It also led in factuality, with a 23% preference rate against Claude 3.7 Sonnet’s 10%.
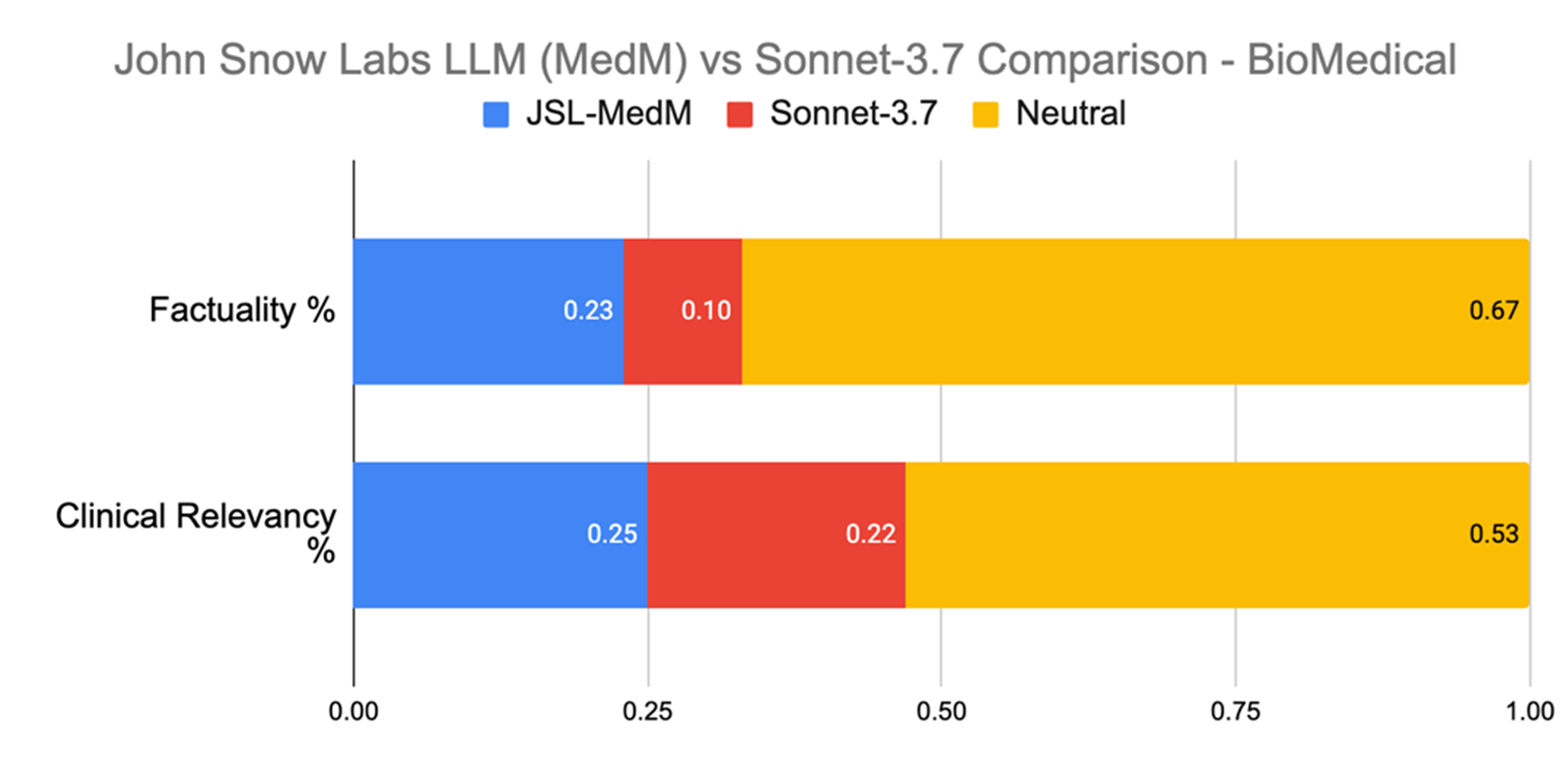
Evaluation on Biomedical Q&A.
These results highlight a key limitation of general-purpose LLMs: while they may be fine for layperson interactions, their lack of clinical alignment and precision can be problematic in high-stakes healthcare settings.
In biomedical Q&A, John Snow Labs’ model was preferred over Claude 3.7 Sonnet more than twice as often for factual accuracy, a dimension crucial for clinical use.
An exception worth noting: structured information extraction
Information extraction remains a foundational task when dealing with unstructured clinical content, involving entity detection, relationship mapping, and linking to standard vocabularies. John Snow Labs brings a decade of experience and a vast catalog of over 2,500 pretrained models available via licensing.
While general LLMs are easy to prototype with, their cost, latency, and inconsistency make them impractical at scale. Extracting three simple facts from 100 million patient notes could cost around $1 million and take several days using GPT-4.5.
Even powerful open models like LLaMA 3.5, with tuned prompts and few-shot learning, struggle to exceed 70% F1 score [1]. Legacy methods like CRFs or Clinical BERT still outperform most LLMs on this task.
In a head-to-head benchmark, John Snow Labs’ healthcare-specific language tools produced five to six times fewer errors than Claude 3.7 Sonnet when extracting nine common clinical entities.
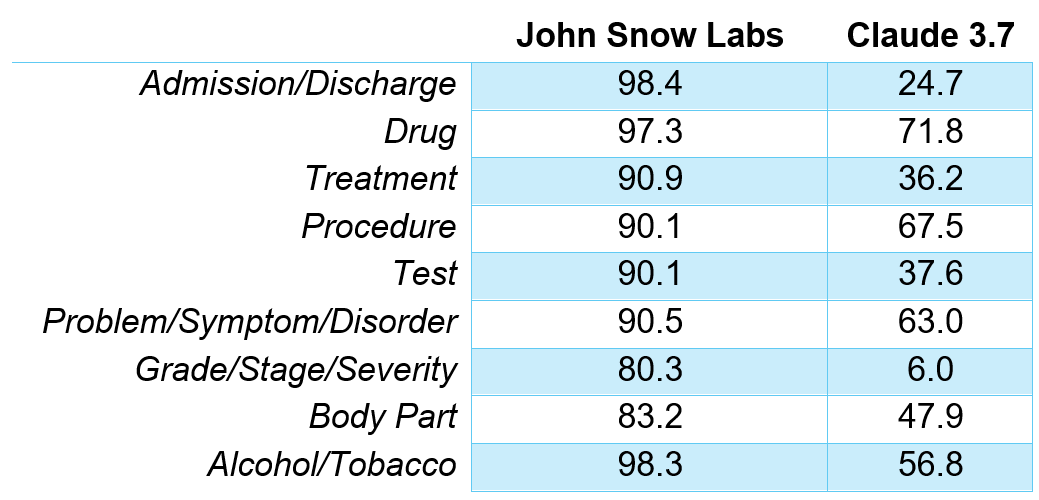
F1 Scores for different entities recognition.
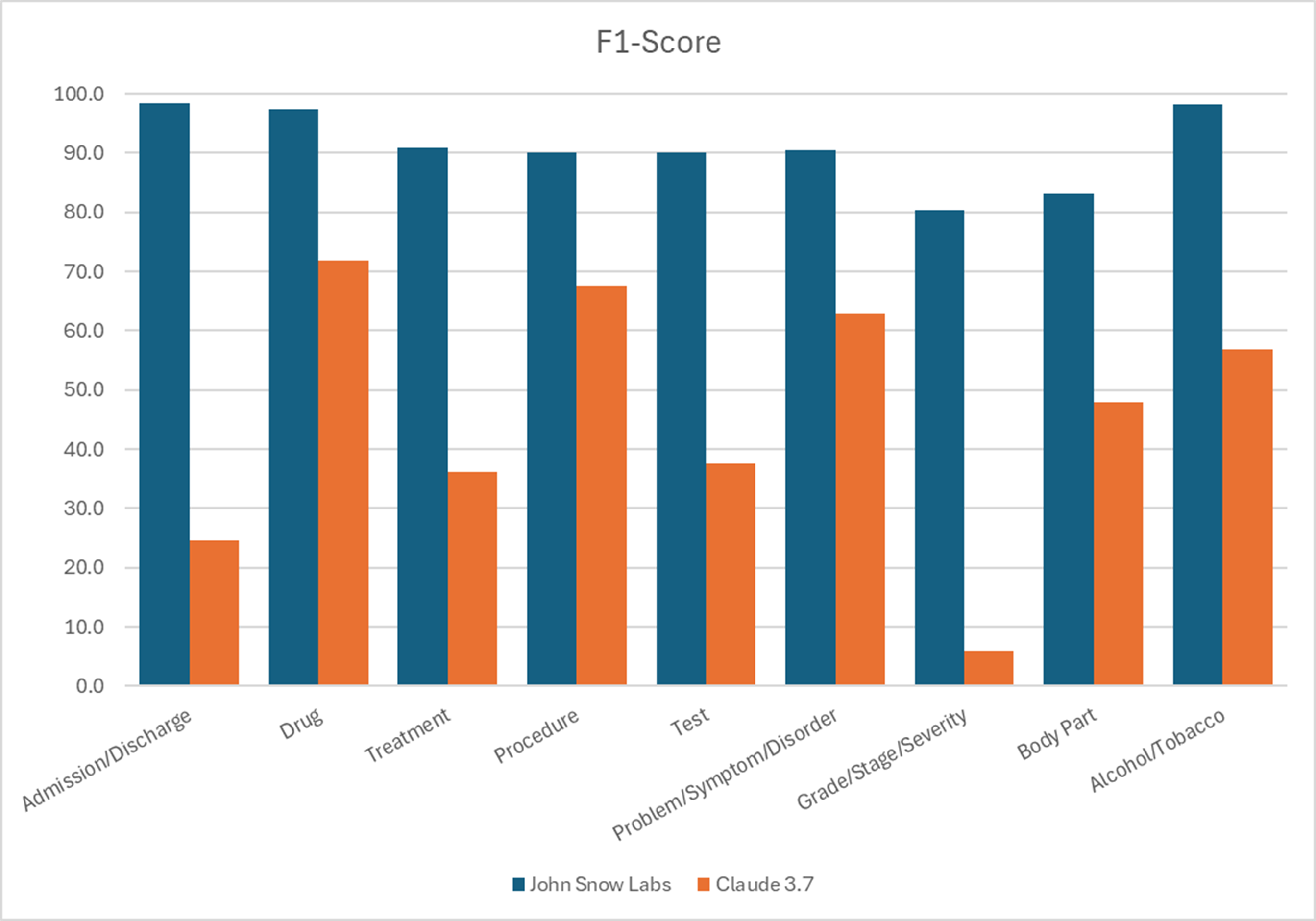
F1 Scores for different entities recognition.
John Snow Labs’ healthcare-specific models made five to six times fewer errors than Claude 3.7 Sonnet in clinical entity extraction.
The suite of John Snow Labs healthcare entity recognition covers more than 400 specific entity types through over 130 dedicated models. These solutions offer a high level of detail including fine-grained categories like 20 disorder subtypes. With easy-to-use visual tools, John Snow Labs’ healthcare pipelines can be built or customized without writing a line of code.
Final thoughts: Weighing performance and operational constraints
Choosing a language model for use in healthcare production environments involves more than reviewing benchmark scores or general capabilities. Although the results discussed here show that models developed specifically for the clinical domain tend to perform better in terms of relevance and factual accuracy—reflected in the consistent preferences shown in blinded evaluations—technical performance is only part of the equation.
Organizations working in healthcare and life sciences operate under strict data protection rules. For many, the ability to run models within their own infrastructure—especially in environments that require isolation, such as air-gapped systems—is not optional but essential. Containerized deployment options and local hosting are not just technical preferences; they often align more closely with legal and institutional requirements for managing sensitive patient data or unpublished research.
Scalability and cost management are also key. Hosted LLMs with usage-based pricing can become cost-prohibitive at scale, particularly in workflows involving large volumes of unstructured clinical text. Alternatives that rely on fixed-cost server licensing may provide better predictability and lower long-term expenses, especially for institutions handling millions of records or operating across multiple sites.
Another relevant point is the pace at which medical knowledge shifts. Tools that receive regular updates—incorporating new datasets, guidelines, or evidence—may better support teams trying to keep up with changes in clinical standards.
John Snow Labs offers secure, on-premise deployment with server-based pricing and continuous updates, making it a valuable choice for the biomedical domain.
Taken together, these aspects suggest that while general-purpose models like GPT-4.5 are versatile and capable, they may not always be the most practical choice for clinical deployments. Models developed with healthcare-specific tasks in mind, and with deployment options suited to secure, high-volume environments, may align more closely with the operational realities and needs of many healthcare organizations.
References
[1] Hsu E, Roberts K. LLM-IE: a python package for biomedical generative information extraction with large language models. JAMIA Open. 2025 Mar 12;8(2):ooaf012. https://pmc.ncbi.nlm.nih.gov/articles/PMC11901043/