Abstract
• Electronic Health Records (EHR) are a rich source of real-world information on opioid drug use that could potentially augment drug safety surveillance at the US Food and DrugAdministration (FDA). Pharmacovigilanceactivities at the FDA are oriented to detect,monitor, characterize, and prevent adverse drug reactions for FDA-approved drugs and therapeutic biologic products. Multiple biomedical resources utilized currently include clinical trials data, spontaneous adverse eventreports, and published scientific literature reports.
• Electronic health records (EHR) are an important information resource about opioid drug allergies. In a study of drug allergies recorded in EHRs of a tertiary healthcaresystem, opioids were the third most frequently implicated drug class.1 In a Veterans Affairs inpatient study, opioids were the second most frequently reported class for drug allergies.2
Objectives
• Garner drug safety insights from EHRs by leveraging rule-based algorithms and deep learning methods to identify opioid-related adverse drug events (ORADE) in discharge summaries from the Medical Information Mart for Intensive Care (MIMIC-III) database3.
• Map drug name and adverse event term mentions from free text to standardized terminologies and ontologies.
• Develop an interactive visualization dashboard to explore structured and unstructured data from the MIMIC-III EHR data to support drug safety review and research
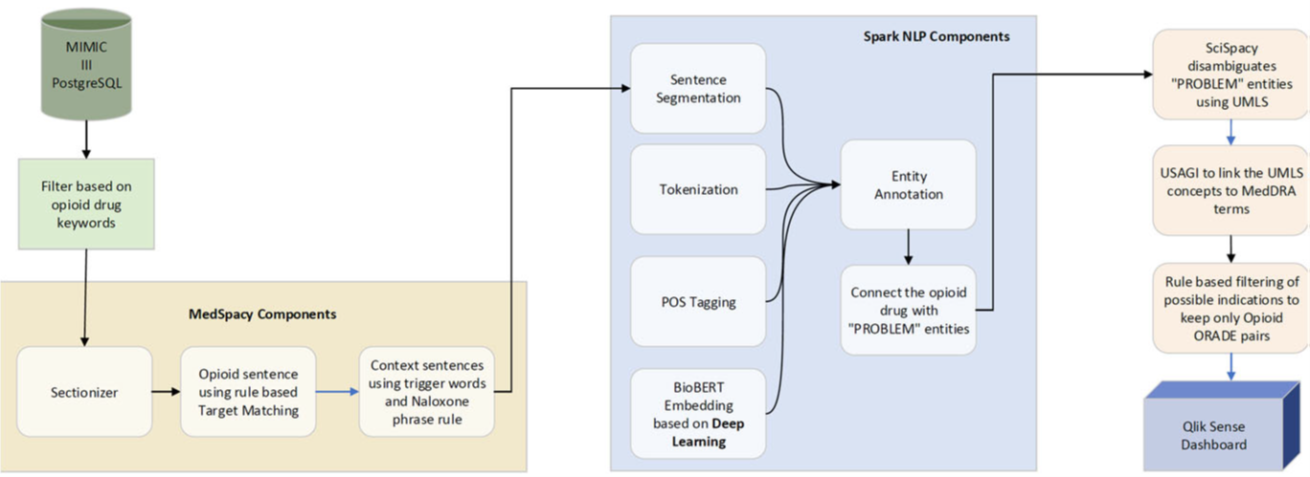
Methods
Sample text from discharge summary highlighted with opioid drug and adverse event mentions in conjunction with trigger phrases to detect potential opioid-related adverse drug event (ORADE) relations:

Hydromorphone – oxygen desaturation Morphine – somnolence
Results
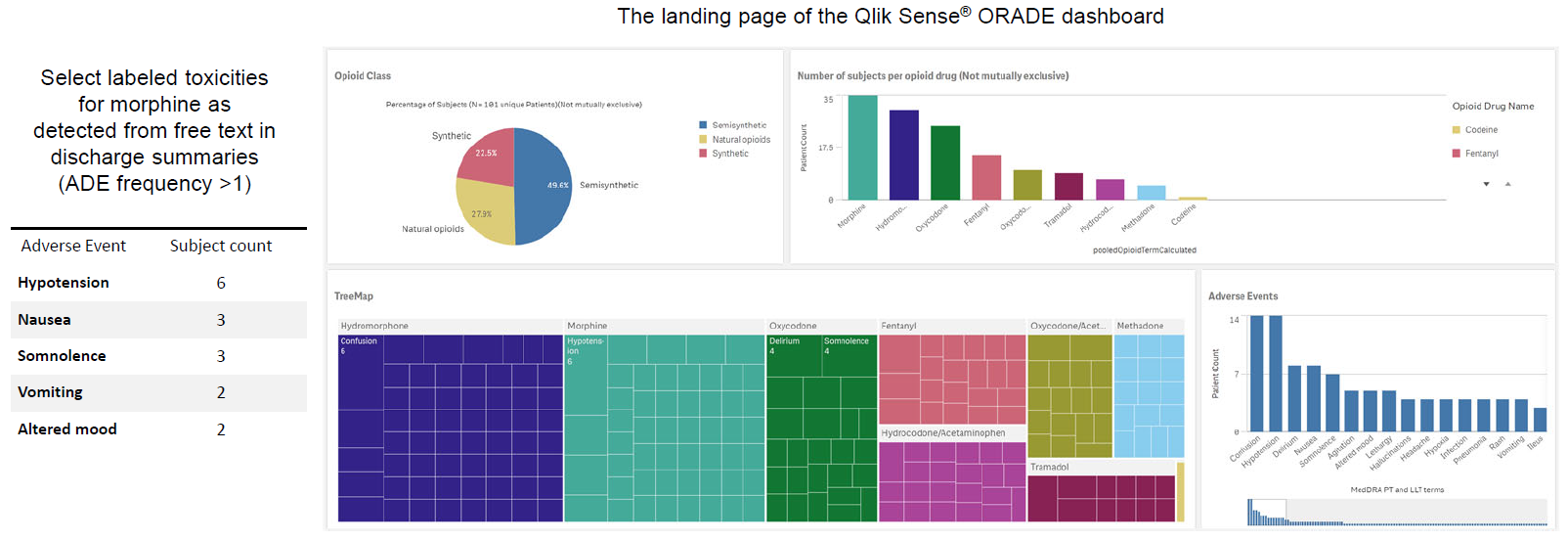
The opioid-ORADE pair detection pipeline was tested on a dataset of 30 opioid-ORADE pairs based on 47 patient’s discharge summaries. This test dataset consisted of the remaining 20% of the originally generated dataset. The trained model achieved accuracy/recall/precision/F1 scores of 0.61/0.6/0.64/0.62, respectively. The extracted candidate drug-event pairs do not indicate causality or absolute risk of an association. Most of the false positives (opioid-ORADE pairs that are detected by the trained model but was not annotated by domain expert) can be attributed to drug regimen change and opioid-indication pairs.
Conclusions and Limitations
CONCLUSIONS
- This novel prototype application provides a practical framework to extract ORADE information from unstructured text in EHR discharge summaries.
ORADE safety signals illustrative of known labeled opioid toxicities can be detected. The application may potentially detect novel emerging ORADEs.
LIMITATIONS
- False positives caused by drug-indication pairs and opioid medication change events
- Concept fragmentation wherein the drug and adverse event do not co-occur in the same sentence
- Complexities in cross-terminology mapping
- Heterogeneity and variable quality of information recorded in the discharge summaries
FUTURE ENHANCEMENTS
Methodological refinements to increase precision in identifying valid candidate ORADE pairs.
Assess the application in detecting new emerging safety issues using a larger and more diversified EHR database
Syed Haque1 (Syed.Haque@fda.hhs.gov), Md Rashedul Hasan2 (MdRashedul.Hasan@fda.hhs.gov), Md Mahmudul Hasan1, Richard Jermyn3,
Ahmad Hussein3, Alex Vega3, Krzysztof Zembrzuski3, Anna Ripple4, Mitra Ahadpour2, Henry Francis2, Alfred Sorbello2
1Research participant at CDER/FDA administered by the Oak Ridge Institute for Science and Education (ORISE); 2US Food and Drug Administration (FDA);
3Rowan University School of Osteopathic Medicine; 4National Library of Medicine/NIH
References
1. Zhou L, Dhopeshwarker N, Blumenthal K, et. al. Drug allergies documented in electronic health records of a large healthcare system. Allergy 2016 Sep; 71(9):1305-13.
2. McConeghy K, Caffrey A, Morrill H, et. al. Are non-allergic drug reactions commonly documented as medication “allergies”? A national cohort of Veterans’ admissions from 2000 to 2014. Pharmacoepidemiol Drug Saf 2017 Apr;26(4):472-476.
3. Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data. 2016 May 24; 3:160035.
Acknowledgements: This project was supported in part by appointment to the research participation program at CDER administered by the Oak Ridge Institute for Science and Education (ORISE) for the FDA and in part by the Intramural Research Program of the National Institutes of Health, National Library of Medicine. Funding support was received from the FDA/CDER/Opioid Research Program.
Disclaimer: The views expressed are those of the authors and do not represent those of the US FDA or the US Government.