Ensuring that electronic health record (EHR) text can be shared without betraying patient privacy is no longer a purely technical exercise. It has become a strategic question of data governance: can we guarantee regulatory-grade protection at scale while preserving the clinical value hidden in free-text?
One-size-fits-all large language models (LLMs) leave too much risk on the table
Independent tests show that even carefully prompted GPT-4 misses almost one in five identifiers in real notes, reaching only 0.83 recall on the 2014 i2b2 reference set [1]. A zero-shot GPT-4o pipeline fares worse, hitting an F1 of 0.79 on a 48-document multisite corpus and driving cloud bills beyond USD 600,000 for an eight-million-note [2]. Such numbers fail the “expert-determination” bar in both HIPAA and GDPR terms, and, more importantly, shatter the audit trail because stochastic sampling means the same file can be de-identified differently on two consecutive runs.
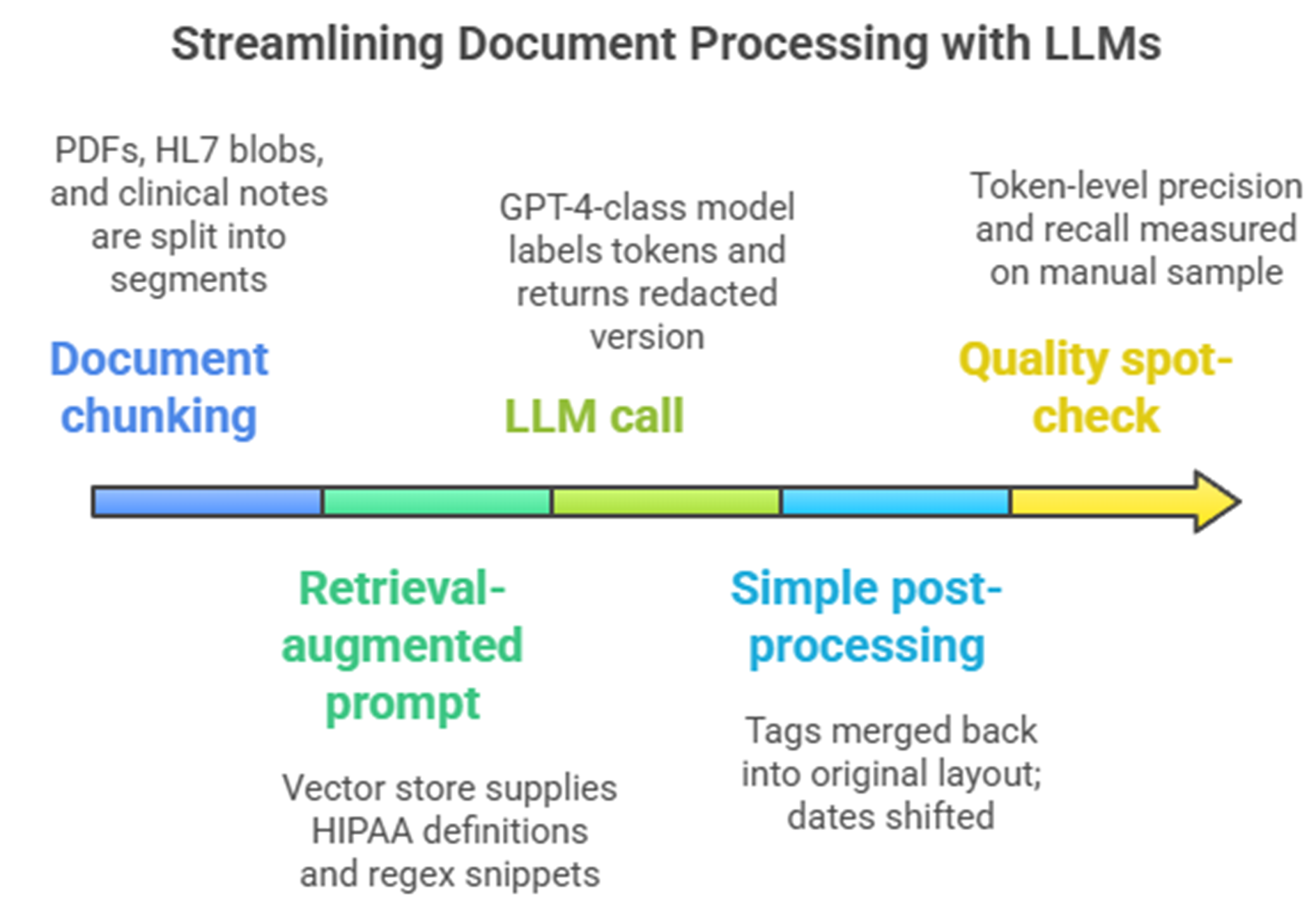
Typical single GPT + RAG de-identification setup
For organizations committed to sharing EHR narratives responsibly, the practical path is neither a human queue nor an all-purpose foundation model, but a purpose-built fusion of medical NER, rule safeguards and cryptographic tagging that turns complexity into confidence.
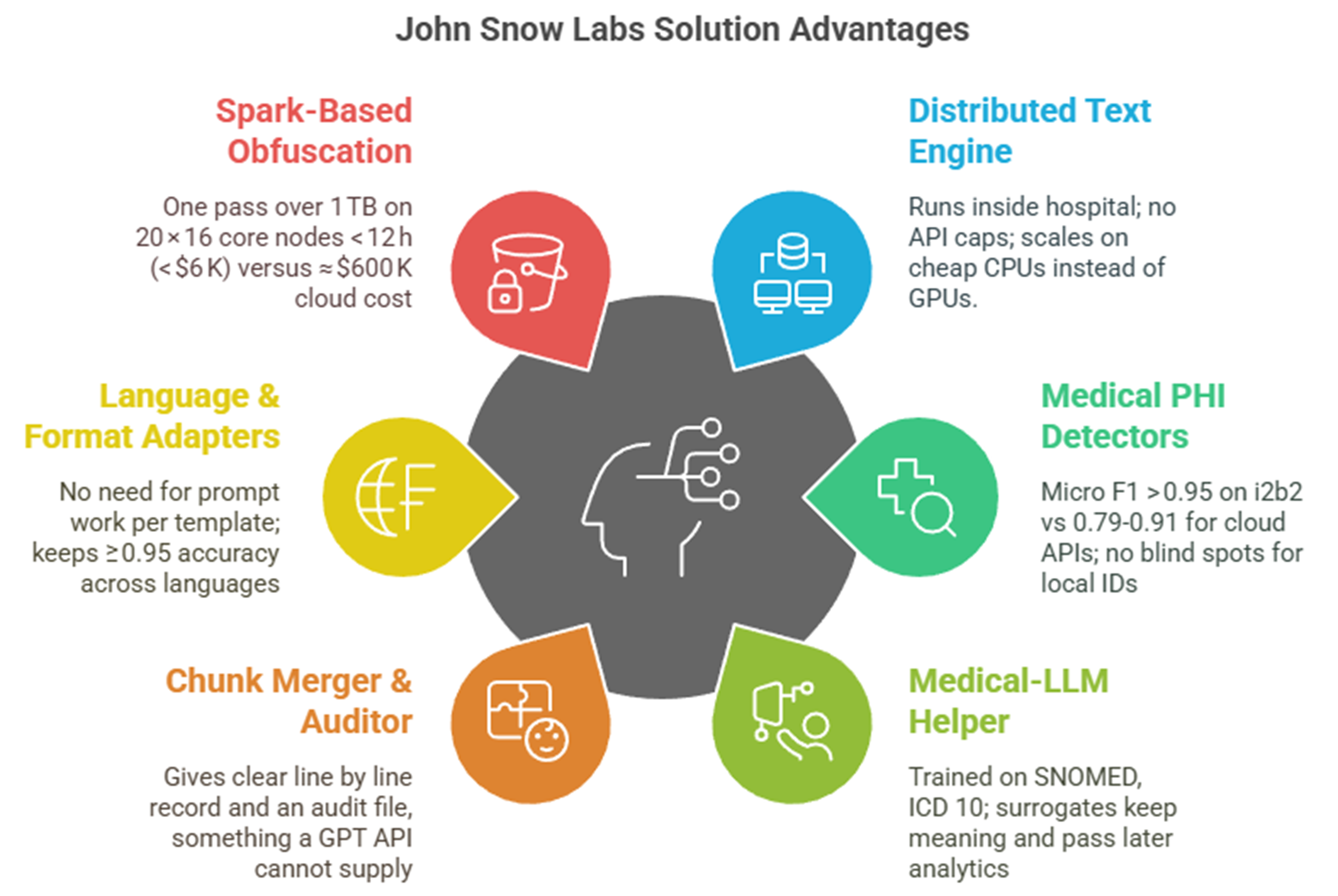
Hybrid, healthcare-native pipelines already meet the 95 %+ recall demanded by regulators at a fraction of the cost.
A production system built on Healthcare NLP blends deep-learning NER, language-aware rules, and reversible hashing. It achieves 96–98 % token-level recall on i2b2 while processing 1.2 TB of mixed-language text overnight on twenty commodity servers for about USD 6,000 compute cost. Across seven European languages, it makes 50 % fewer errors than AWS Comprehend Medical, 475 % fewer than Azure, and 33 % fewer than ChatGPT-4 [4]
|
Dimension |
Healthcare-aware stack |
Generic GPT + RAG |
|
PHI recall |
96–98 % |
80–90 % |
|
Cost for 8 M notes |
≈ USD 90 K license + 6 K compute |
500–800 K usage fees |
|
Multilingual |
Seven languages out-of-the-box |
Prompt engineering needed; recall drops |
|
Auditability |
Deterministic, per-token confidence, SHA-256 re-hash |
Non-repeatable, probability opaque |
De-identification is a moving target: governance teams need determinism, surrogate consistency, and language coverage
Rule-only systems falter on nuances such as Spanish phone numbers or French social-security IDs, while general LLMs hallucinate replacement values that break longitudinal links. The Healthcare NLP approach keeps locale-aware fake values stable across documents, attaches reversible hashes for ethics-board re-identification and integrates new regex guards in minutes, shielding teams from “template drift” as note styles evolve.
Taken together, these findings redraw the risk-benefit map for secondary use of EHR text. Governance leaders no longer have to choose between manual redaction or black-box generative AI. A domain-tuned, hybrid pipeline delivers regulatory-grade privacy, predictable economics and verifiable audit trails, clearing the last operational hurdle to transform clinical free-text into reproducible real-world evidence.
Generative AI Lab: De-identification with human expert review healthcare
For non-data scientists, the out-of-the-box de-identification capabilities in John Snow Labs’ Generative AI Lab on AWS Marketplace is the way to go. It not only makes the review and approval (human in the loop) process easier, the feedback can also allow subject matter experts to improve the accuracy as the models learn.
The workflow for de-identification using Generative AI Lab begins with the secure storage of clinical data, such as patient notes, lab reports, and medical images in customers’ own AWS environment. John Snow Labs NLP models then analyze these unstructured documents to identify PHI information. This identified PHI information can then be configured to perform masking or obfuscation. While Generative AI Lab handles much of the automation, external systems like cloud storage, EHRs, and patient demographic databases are essential for data input ensuring seamless integration and compliance with healthcare regulations.
Key takeaway
[1] B. Altalla’ et al., “Evaluating GPT models for clinical note de-identification,” Sci Rep, vol. 15, no. 1, p. 3852, Jan. 2025, doi: 10.1038/s41598-025-86890-3.
[2] V. Kocaman, M. Santas, Y. Gul, M. Butgul, and D. Talby, “Can Zero-Shot Commercial APIs Deliver Regulatory-Grade Clinical Text DeIdentification?,” Mar. 31, 2025, arXiv: arXiv:2503.20794. doi: 10.48550/arXiv.2503.20794.
[3] V. Kocaman, H. U. Haq, and D. Talby, “Beyond Accuracy: Automated De-Identification of Large Real-World Clinical Text Datasets,” 2023, doi: 10.48550/ARXIV.2312.08495.