De-Identifying Whole Slide Images (WSI) on AWS SageMaker: Complete Deployment Walkthrough — Part 3
Protected health information (PHI) is often embedded in SVS files used by healthcare professionals. As explained in Part 1, this PHI must be removed before sharing or analyzing data for research, AI training, or diagnostics.
Here’s how to quickly set up SVS DEID on AWS SageMaker in just a few minutes:
Step 1 – Import the listing from AWS Marketplace
1.1 Visit the SVS Images De-identification listing on AWS Marketplace, and click Continue to Subscribe.
1.2 Review and accept the EULA, pricing, and support terms.
1.3 Click Continue to Configuration, choose a region, and copy the Product ARN shown. You’ll use it when creating a deployable model with Boto3.
Step 2 – Deploy the imported model as endpoint
Choose your instance type based on your performance and cost preferences. You can refer to the provided benchmarks for guidance.
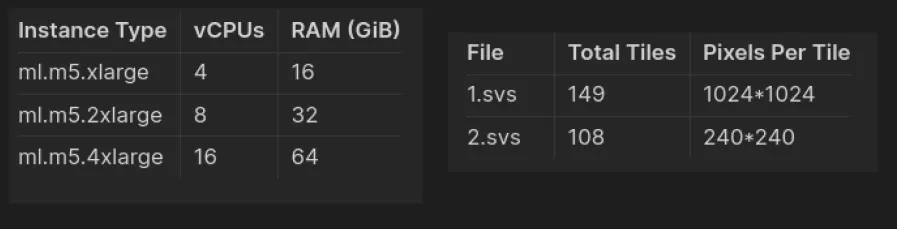
Benchmarked Instances and files
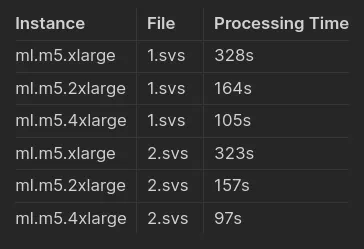
Benchmark Results
While deploying make sure to:
- Select async configuration and provide the S3 path where DEID output files will be stored.
- SageMaker role has read/write access to the output S3 folder, and read access to an input folder
Step 3 – Upload SVS Files and Test the De-identification Endpoint
3.1 Upload input SVS file to S3
# Step 1: Download the SVS file from GitHub
svs_url = "https://raw.githubusercontent.com/JohnSnowLabs/visual-nlp-workshop/refs/heads/master/emr/input3.svs"
file_name = svs_url.split('/')[-1]
response = requests.get(svs_url)
with open(local_path, "wb") as f:
f.write(response.content)
# Step 2: Upload to S3
s3_client.upload_file(file_name, "svs-sage-input", file_name)
print(f"Uploaded {file} to s3://{bucket_name}/{s3_key}")
3.2 Deploy Model with model ARN
model_package_arn = 'arn:aws:sagemaker:YOUR ARN FROM STEP 1'
model = ModelPackage(
role=role,
model_package_arn=model_package_arn,
sagemaker_session=session,
)
model.deploy(
initial_instance_count=1,
instance_type='ml.m5.2xlarge',
endpoint_name='svsdeid',
async_inference_config=AsyncInferenceConfig(
output_path="s3://svs-sage-output/outputs/"
)
)
model.endpoint_name
3.3 Call the endpoint with s3 SVS file path
sm_runtime = boto_session.client("sagemaker-runtime", region_name='us-east-1')
response = sm_runtime.invoke_endpoint_async(
EndpointName='svsdeid',
ContentType='application/octet-stream',
InputLocation='s3://svs-sage-input/input1.svs',
Accept='application/octet-stream',
)
response
Optional: Specify tags to remove:
deid_tags = [
'ImageDescription.ScanScope ID',
'ImageDescription.Time Zone',
'ImageDescription.ScannerType',
]
custom_attributes = f"svs_tags={','.join(deid_tags)}"
response = sm_runtime.invoke_endpoint_async(
EndpointName=endpoint_name,
ContentType='application/octet-stream',
InputLocation=input_s3_path,
Accept='application/octet-stream',
CustomAttributes=custom_attributes
)
While its processing you can check the endpoint logs in the Sagemaker Endpoint UI if the permissions are fine and the processing has started.
The response will look like
{
"OutputLocation": "s3://svs-sage-output/outputs/uuid.out",
...
}
You can find more indepth Python tutorial in this notebook
Note: Only async processing is supported. Synchronous (invoke_endpoint) calls will timeout.
Conclusion: Secure and Scalable De-Identification for WSI on SageMaker
AWS SageMaker offers a secure, scalable solution for de-identifying Whole Slide Images (WSI) containing protected health information. By combining SVS-specific metadata removal with asynchronous inference, this deployment minimizes risk while maximizing throughput. Whether you’re managing clinical pipelines or preparing data for AI development, this walkthrough helps you implement an efficient, compliant workflow in minutes. With built-in customization and clear benchmarking, the process empowers healthcare teams to uphold data privacy without sacrificing speed or accuracy.
FAQ
What is the purpose of deploying SVS de-identification on SageMaker?
Deploying the Visual NLP SVS de-identification pipeline on SageMaker enables scalable, automated removal of PHI from Whole Slide Images in a secure cloud environment.
How do I get started with the SVS DEID model on AWS?
Begin by subscribing to the SVS De-identification model on AWS Marketplace, accepting the terms, and copying the model’s ARN to deploy with Boto3.
Which instance type should I choose for deployment?
You can select an instance type based on your budget and performance needs. Benchmark results are available to guide your choice.
Can I customize which tags are removed during de-identification?
Yes, optional metadata tags such as ScanScope ID, Time Zone, and ScannerType can be specified using the CustomAttributes field when invoking the endpoint.
What type of inference is supported for SVS de-identification?
Only asynchronous inference is supported (invoke_endpoint_async). Synchronous calls will timeout due to the size and processing time of SVS files.
Where is the output stored after de-identification?
The de-identified SVS files are saved to a specified S3 output folder. Make sure the SageMaker role has the appropriate permissions for S3 access.
Is there a Python example available to follow?
Yes, a complete Python tutorial is available as a Jupyter notebook, showing how to upload input files, deploy the model, and call the endpoint.
Who should deploy this pipeline?
This solution is ideal for healthcare data engineers, clinical AI developers, and researchers who need to process large volumes of medical images securely and compliantly in the cloud.
Understand Visual Documents with High-Accuracy OCR, Form Summarization, Table Extraction, PDF Parsing, and more.
Learn More