What to Know Before De-identifying Whole Slide Images (WSI)
Discover the low-level, hands-on steps of de-identifying SVS and DICOM medical images using Visual NLP, focusing on PHI hidden in metadata and pixels. Part 1 of a three-part series for privacy-focused healthcare AI workflows.
This is the first post in a three-part series on how to de-identify Whole Slide Imaging (WSI) files using the Visual NLP library by John Snow Labs. We’ll focus on two common formats in digital pathology: SVS and DICOM.
What You’ll Learn About SVS and DICOM De-Identification
- What Visual NLP is and how it fits into medical imaging workflows.
- A brief introduction to the SVS and DICOM formats, how WSI data is structured, and where sensitive information can hide.
- Types of Protected Health Information (PHI) that may be embedded in metadata or pixel data.
- Why de-identification is essential for privacy, compliance, and collaboration.
What’s coming in Part 2?
- A step-by-step breakdown of the de-identification pipeline using the Visual NLP library from John Snow Labs.
- What the output looks like: a clean SVS or DICOM file, ready for secure use.
What’s coming in Part 3?
- Turning the pipeline into a cloud-based service with an API.
- Deploying the system on AWS SageMaker.
📝 Note: The first two posts focus on the low-level, hands-on steps of de-identifying medical images, while the final post moves to a high-level, cloud-based perspective, showing how everything fits together in production.
De-Identification at scale
Hospitals and research centers are collecting more and more medical images. To protect patient privacy, this data needs to be de-identified. Doing it manually takes a lot of time, can lead to mistakes, and doesn’t work well when there are thousands of images.
This is where Visual NLP steps in.
How Visual NLP Automates Medical Image De-Identification
Visual NLP is a library built on Apache Spark, designed for Visual Document Understanding and OCR (Optical Character Recognition) tasks, including SVS and DICOM de-identification, at scale. Here’s how it can revolutionize the process:
Scalability: Processes large DICOM datasets in parallel using Apache Spark.
Automation: Removes sensitive data without manual effort.
Customization: Supports custom rules for specific privacy needs.
Accuracy: Uses OCR to precisely detect and remove text.
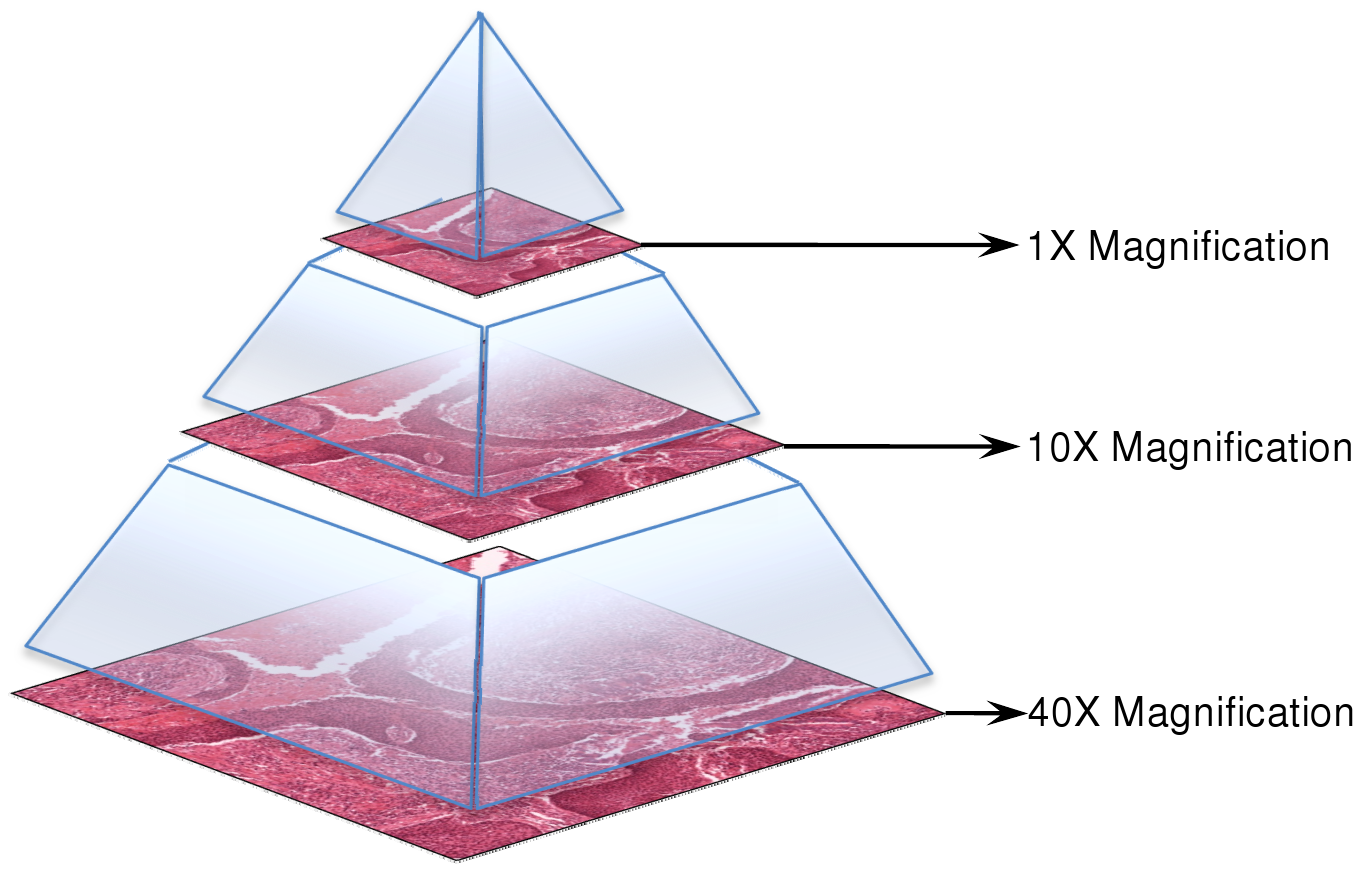
SVS Format: Where PHI Can Hide

How it looks like an svs file in Aperio
SVS Structure:
- A pyramid of images at various zoom levels (e.g., 40x, 20x).

Image from nema.org
- Each level contains tiles, not full images — ideal for parallel processing.
Where PHI May Be Embedded in SVS files?
- Label images (e.g., scanned patient ID stickers).
- Macro images (overview shots with annotations).
- Metadata: custom tags may include patient names, dates, institutions, scan details.
- Burned-in pixel text (e.g., names or IDs printed on the slide).
Understanding DICOM Format and Where PHI Resides
DICOM is the standard for medical imaging — used in everything from MRIs to digital pathology.
PHI in DICOM Can Appear In:
- Metadata: Standard tags like patient name, ID, birth date, institution; may also include private/vendor-specific fields.
- Pixel data: Burned-in text embedded directly in image pixels.
- Overlay data: Annotations or graphics overlaid on images.
- Encapsulated documents: Embedded PDFs, text, or HTML files.
Why De-Identifying SVS and DICOM Files Is Essential for Compliance
It’s more than a best practice — it’s a legal and ethical obligation.
✅ Stay Compliant: Laws like HIPAA (U.S.) and GDPR (EU) require removing PHI before sharing or secondary use.
🤝 Enable Safe Collaboration: Researchers, hospitals, and companies can collaborate freely using de-identified datasets.
🤖 Train AI Responsibly: PHI-free datasets are essential for developing trustworthy AI in digital pathology.
🏥 Build Trust: Strong privacy practices improve confidence among patients, providers, and researchers.
Summary: Key Takeaways on De-Identifying SVS and DICOM Files
This post covered the essentials of de-identifying SVS and DICOM WSI files, where PHI hides, how it’s stored, and why it matters. From label images and metadata in SVS files to burned-in text and overlays in DICOM, sensitive information can appear in both expected and hidden places. Understanding these structures is the first step toward building compliant, privacy-preserving workflows that enable research, collaboration, and responsible AI development in digital pathology.
🔜 What’s Next?
In Part 2, we’ll show how Visual NLP helps automate this process.
Resources
- Aperio ImageScope
- WSI Documentation
- WSI Structure Doc
- DICOM Standard Documentation
- DICOM WSI
- Visual NLP
FAQ:
What is Visual NLP and how is it used in medical imaging?
Visual NLP is a library from John Snow Labs designed for Visual Document Understanding and OCR tasks. In medical imaging, it automates the de-identification of WSI formats like SVS and DICOM to protect sensitive data.
Why is de-identification important for SVS and DICOM files?
De-identification removes Protected Health Information (PHI) to ensure compliance with privacy laws like HIPAA and GDPR. It also enables safe collaboration and AI model training.
What types of PHI can be found in WSI files?
PHI may appear in metadata (e.g., patient names, scan dates), label and macro images, burned-in pixel text, and even encapsulated documents in DICOM files.
How is PHI embedded in SVS and DICOM formats?
In SVS files, PHI may be hidden in label images, metadata, or image tiles. DICOM files may contain PHI in standard metadata fields, burned-in text, overlays, or embedded documents.
What formats does Visual NLP support for de-identification?
Visual NLP supports SVS and DICOM formats, offering scalable, automated de-identification for both.
Who should read this article?
This article is ideal for healthcare IT professionals, AI developers, and researchers working with digital pathology data who need to ensure privacy compliance at scale.