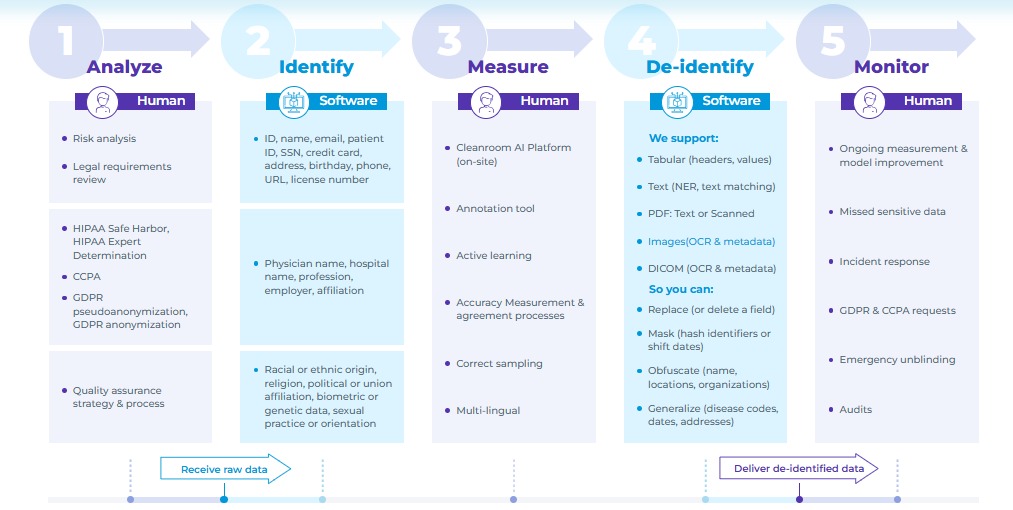
Protected health information (PHI) has become both a strategic asset and a growing liability. In 2024, healthcare data breaches cost organizations an average of $9.77 million, positioning healthcare as the most expensive industry for security failures. At the same time, regulators are stepping up enforcement: the Office for Civil Rights issued $144.9 million in penalties across 152 cases, holding organizations directly accountable for the exposure of PHI.
These risks are amplified by the current operational culture of many healthcare teams. Remote reviewers using personal or unmanaged devices introduce new vulnerabilities that are often beyond the reach of traditional security policies.
Meanwhile, the rapid adoption of AI has introduced a flood of unstructured multi-modal data that must be inspected and approved before it can be used safely in model development or shared with partners.
The combination of rising breach costs, stricter penalties, remote access risks, and growing data volumes points to one bottleneck: human-in-the-loop (HITL) reviewers can’t scale their work or share results safely until PHI is fully masked and verified.
This article demonstrates how no-code de-identification can help you transform this bottleneck into a competitive advantage, empowering your teams to review data more efficiently without compromising compliance.
De‑Identification: The Foundation of Safe HITL
Removing PHI at the very start of the workflow transforms the review from a choke point into a low‑friction routine. The following sections offer a roadmap to a robust HITL de-identification process.
Broadened Access Starts with Removing Every Identifier
The Health Insurance Portability and Accountability Act (HIPAA) outlines eighteen identifiers that classify a record as protected health information. But if even one of these elements stays in the record, it’s still considered PHI. It brings the full weight of HIPAA compliance into play and limits how your team can access or review it.
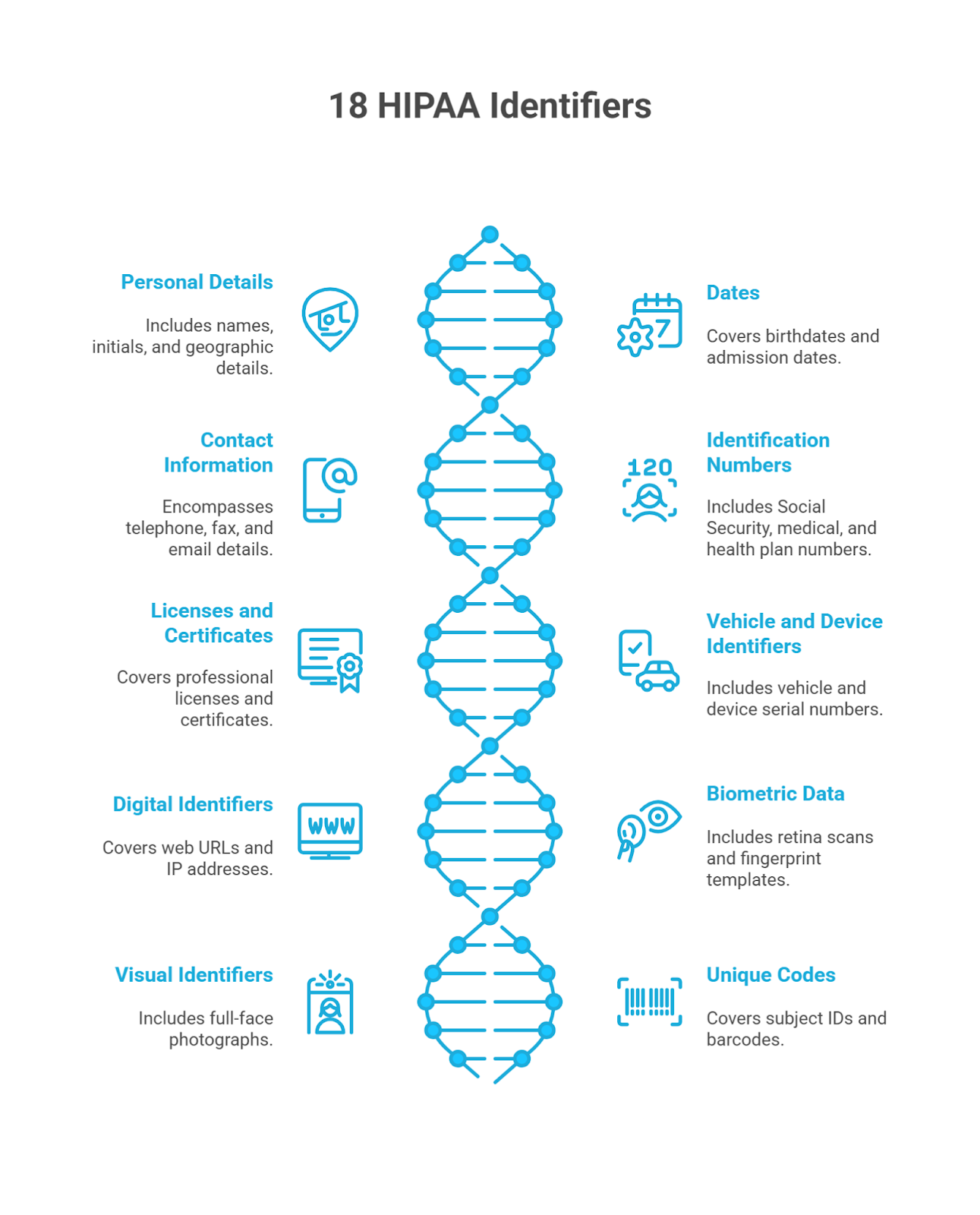
Any one of these 18 identifiers in proprietary data brings HIPAA rules, access limits, and audit requirements into play.
No-code de-identification solves this challenge by removing or masking these elements as soon as the data enters your pipeline. Masking deletes the identifier entirely, while obfuscation replaces it with a realistic surrogate such as a shifted date or an alternate name. Once all PHI is removed from the data, reviewers can access data securely using standard devices and workflows.
De-identification of patient data for research and compliance in Generative AI Lab.
Security teams no longer need to maintain isolated review environments, while legal and compliance workflows become more streamlined without the burden of additional consent protocols. As a result, teams can move faster with greater confidence, knowing the data they’re working with is already safe to use.
Simplifying Compliance Starts with an Immutable Audit Trail
Regulators expect you to prove, step by step, how every piece of PHI was handled. Traditional methods, such as email threads, shared drives, or spreadsheet trackers, often leave gaps that auditors can question, and your legal team must patch them under pressure.
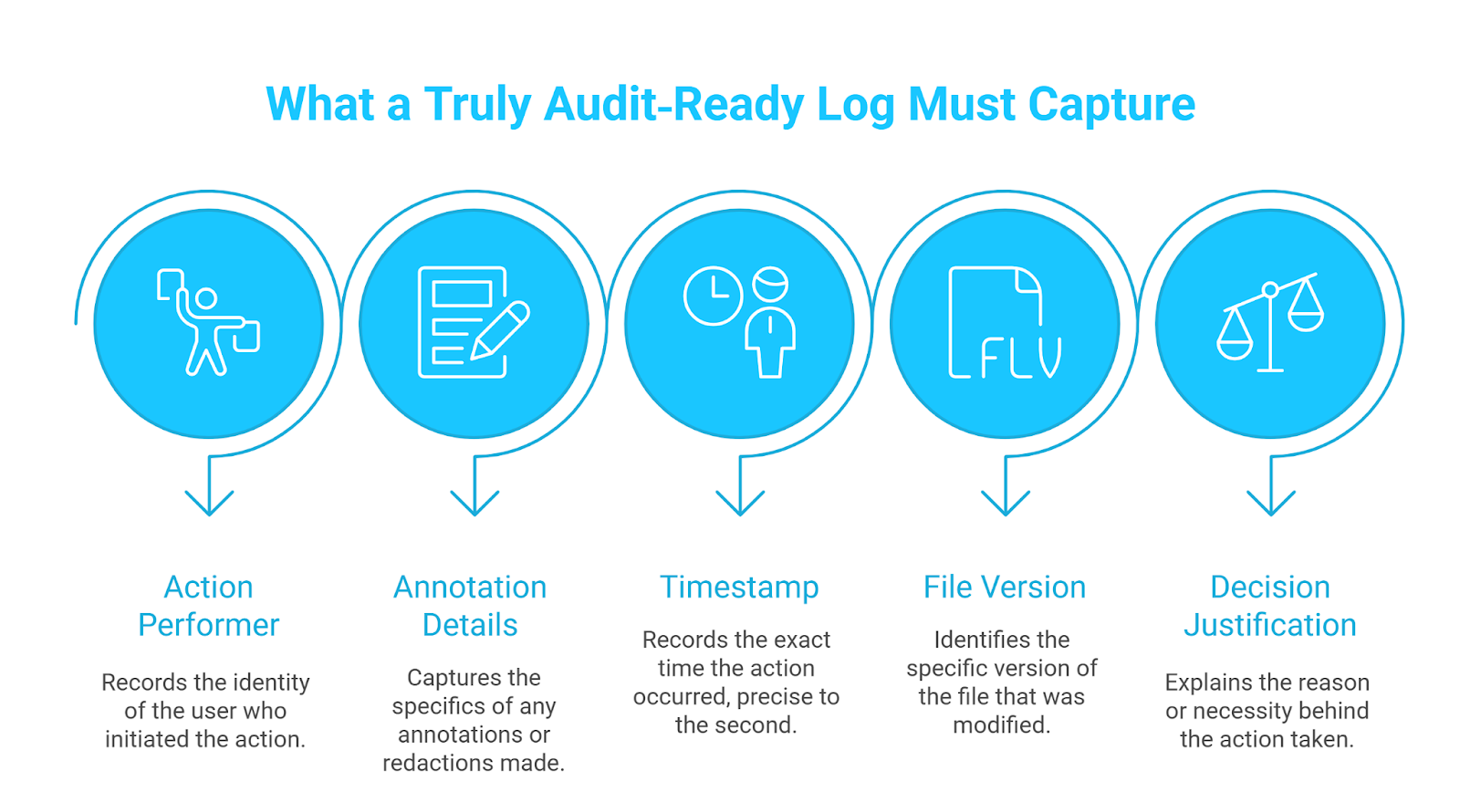
Audit-ready logs track who did what, when, where, and why to each record.
With no-code de-identification, each annotation and redaction is written into an append-only log the moment it occurs. Teams can open the entire decision chain with a single click. Auditors no longer have to chase email chains, compliance teams can avoid last‑minute document hunts, and your organization can prove due diligence in minutes instead of days.
Scaling Review Workflows Without Compliance Bottlenecks
Large AI-based healthcare projects often slow to a crawl because every new reviewer, partner, or business unit triggers another round of consent forms and security reviews. The bottlenecks usually appear as:
- Lengthy legal reviews for every data-sharing agreement
- Vendor risk assessments that stretch project timelines
- Limited seats in secure “clean rooms” that cap team size
No-code de-identification eliminates these friction points by removing PHI at the start, so the data no longer falls under strict privacy regulations. As a result, your team can bring in reviewers and external partners without triggering new compliance protocols or waiting through weeks of legal reviews. Distributed teams can begin work within hours, and review cycles can move faster.
Scale review workflows without compliance bottlenecks.
Generative AI Lab’s No-Code HITL Workflow
Building an in-house de-identification pipeline typically involves recruiting engineers, training clinicians on multiple tools, and writing code for each data type. Generative AI Lab replaces that complexity with a single browser workspace where teams review, validate, and release data without writing code.
Centralize Text Data Within a Single Workspace
Import clinical text such as discharge summaries, clinic notes, and chat transcripts into one unified workspace. The text de-identification template allows you to configure masking rules, define surrogate replacements, and export anonymized outputs without writing code. This ensures consistent handling of PHI across all files and simplifies review workflows for your team.
AI-powered pre-labeling to kickstart medical data annotation in Gen AI Lab.
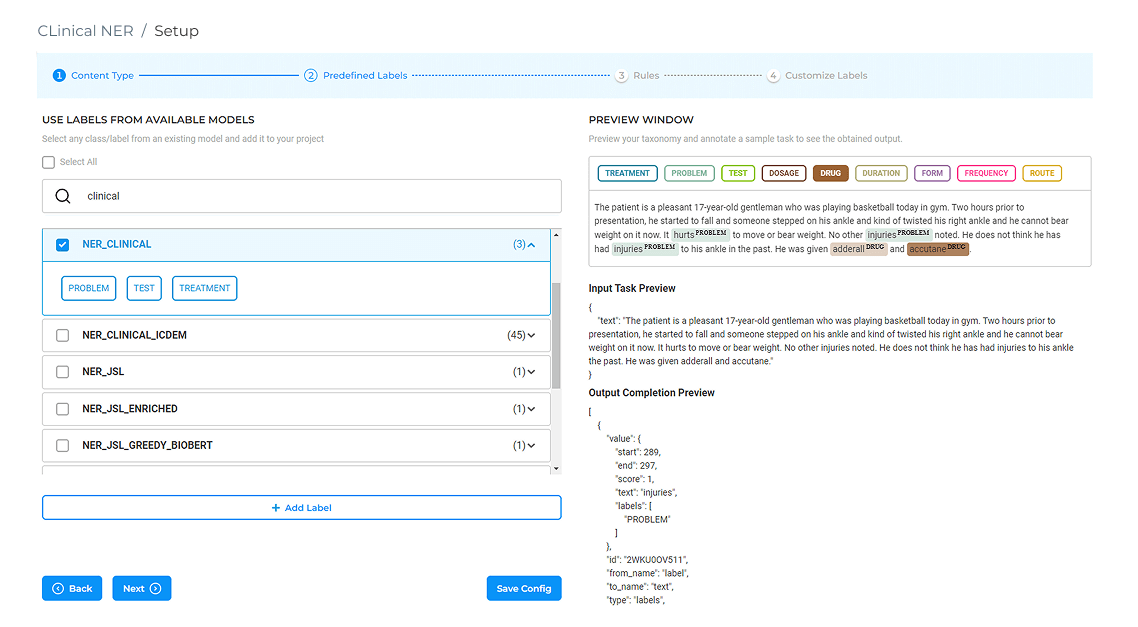
AI-Powered Annotation with 6,600+ NER Models
More than 6,600 Healthcare NLP models can label PHI, clinical terms, and specialty‑specific entities before a human opens the file. Domain experts begin with pre-tagged drafts, shifting their effort from initial markup to quick verification while server safeguards maintain predictable performance.
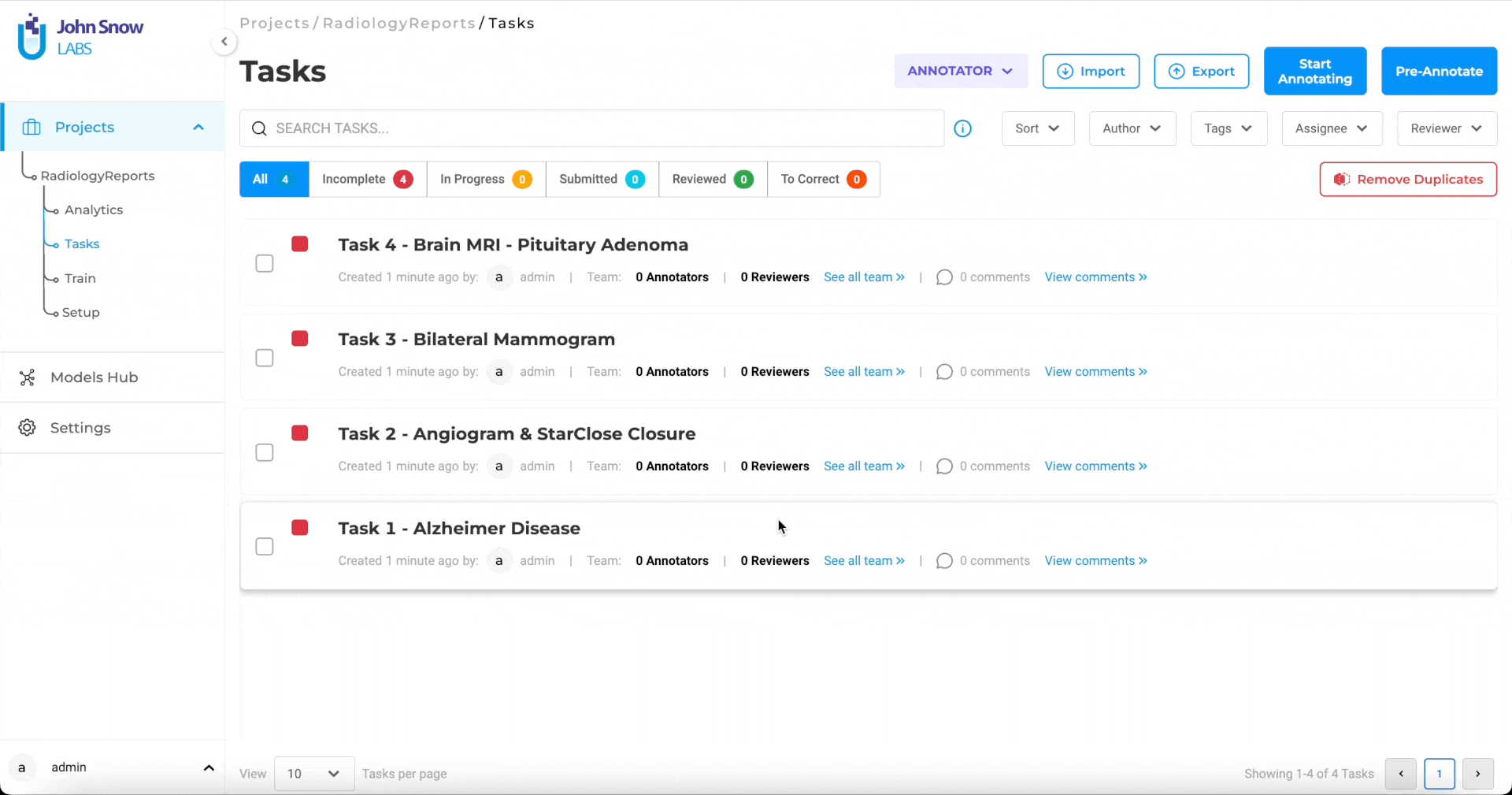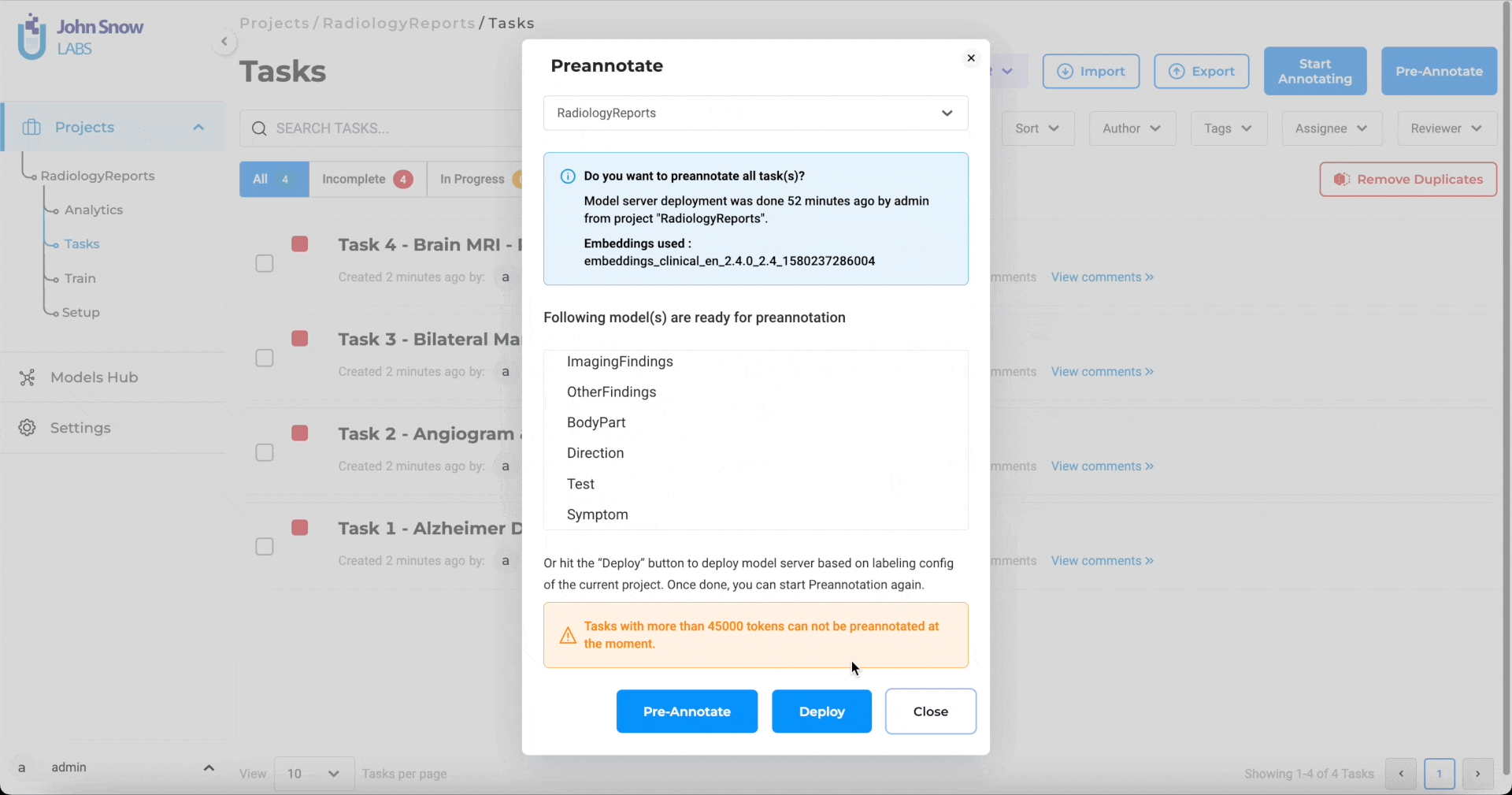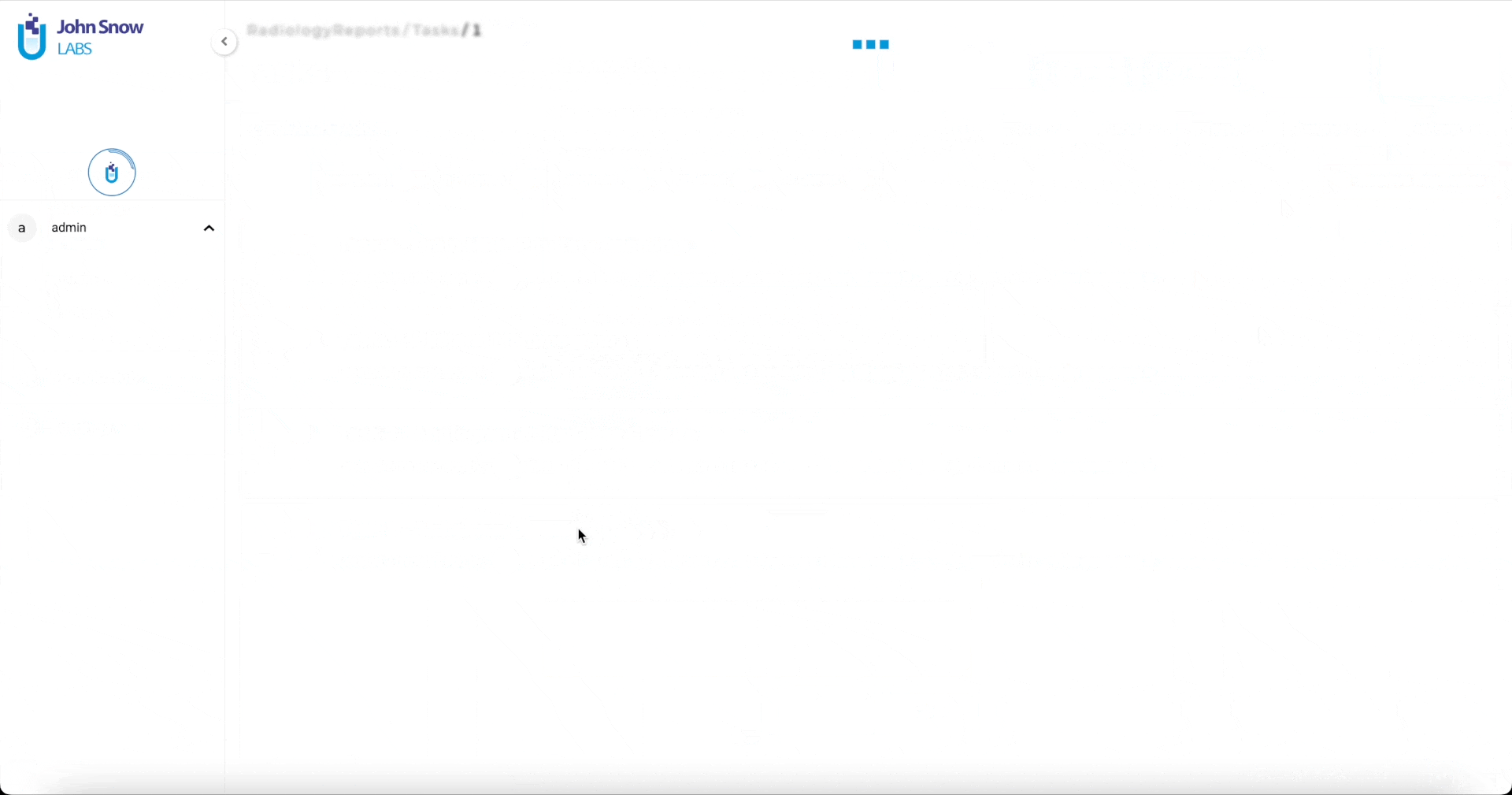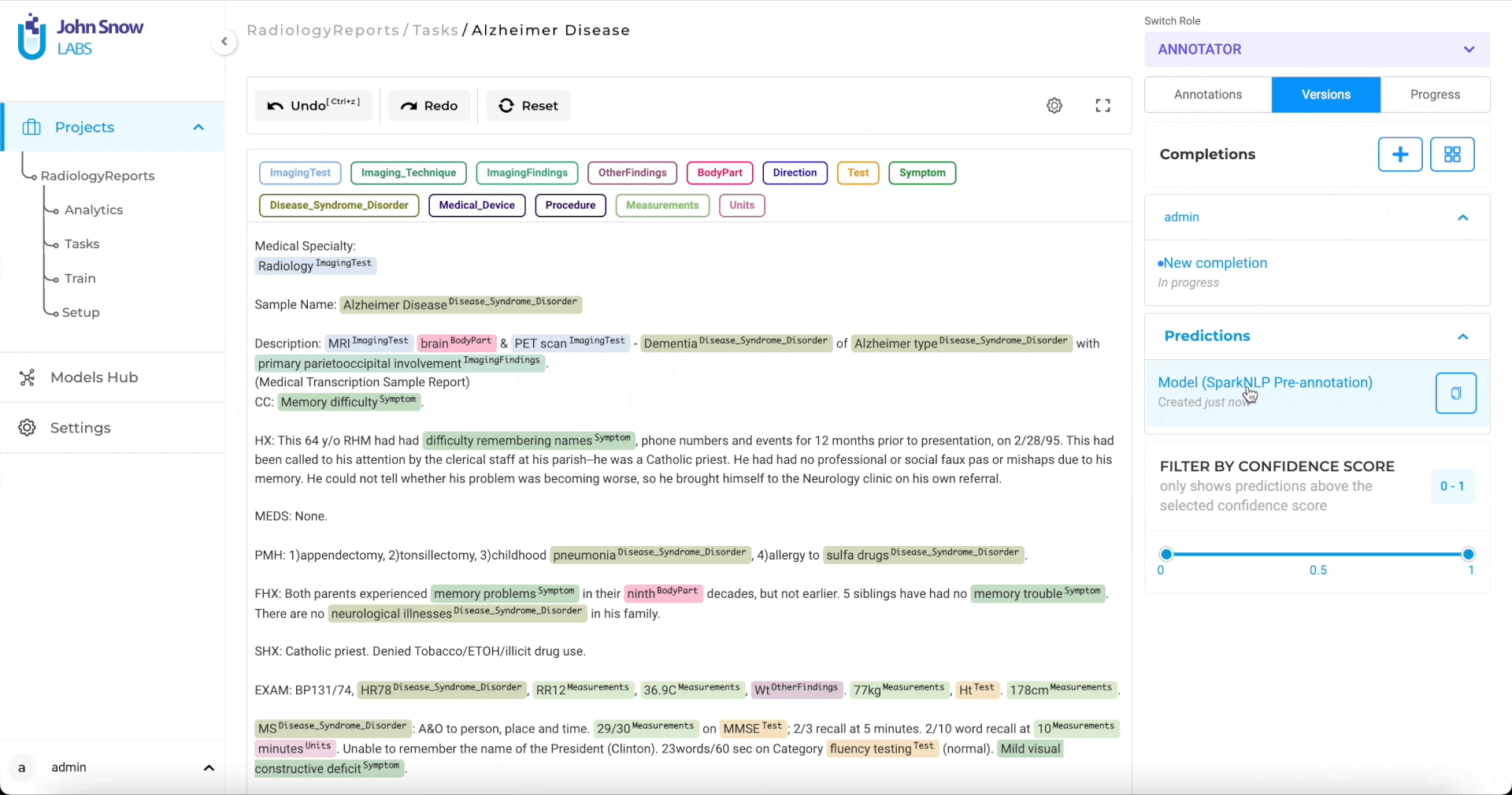
Pre-annotation of patient records using 6,600+ Healthcare NLP models.
Distribute Workload Efficiently with Bulk Assignment
Assign up to 200 documents at once to selected team members. You can choose to assign tasks in sequence or randomly, depending on the project’s needs. This eliminates the need for manual tracking, keeps reviewers productive, and helps maintain a balanced workload across the team.
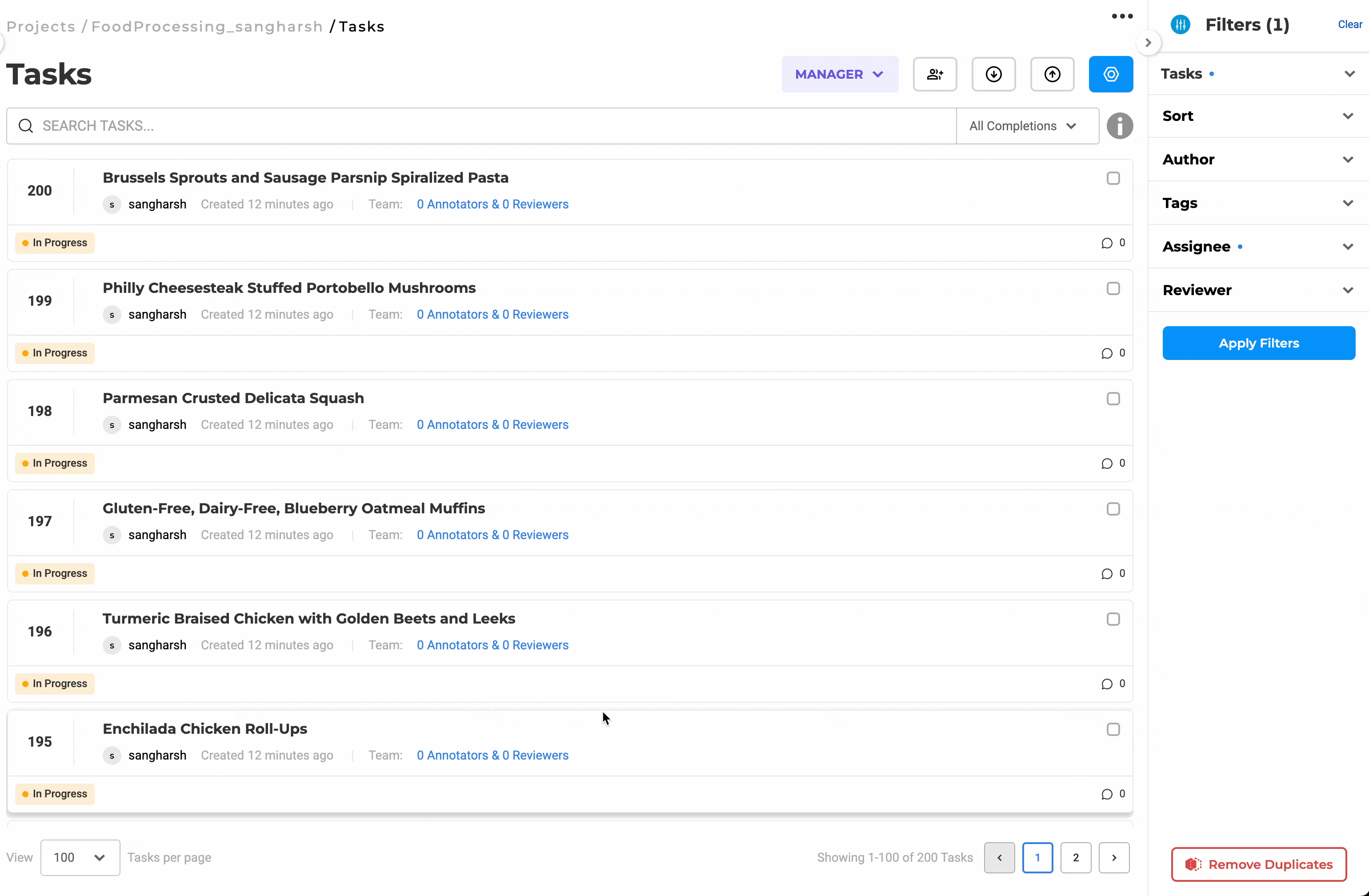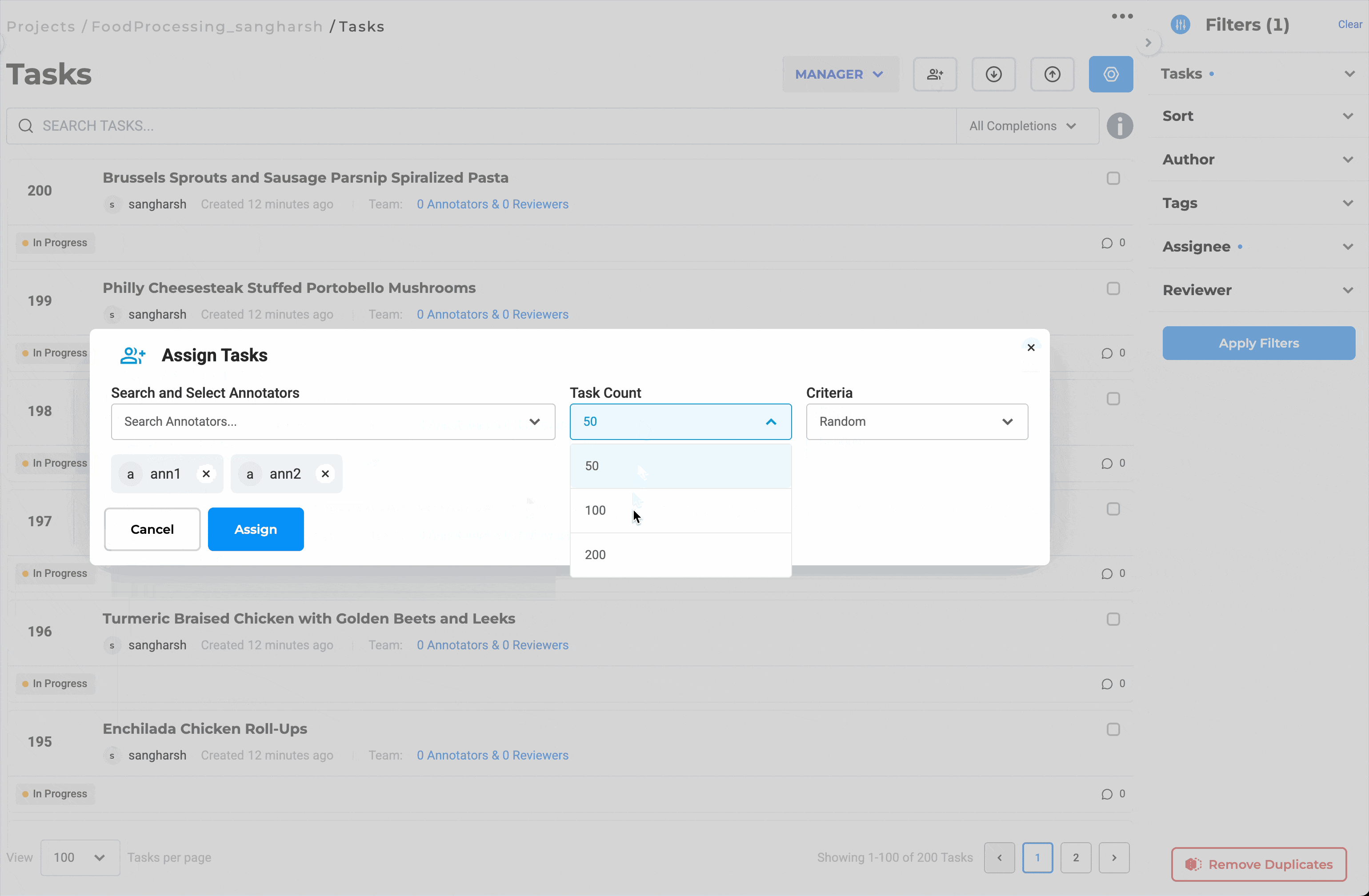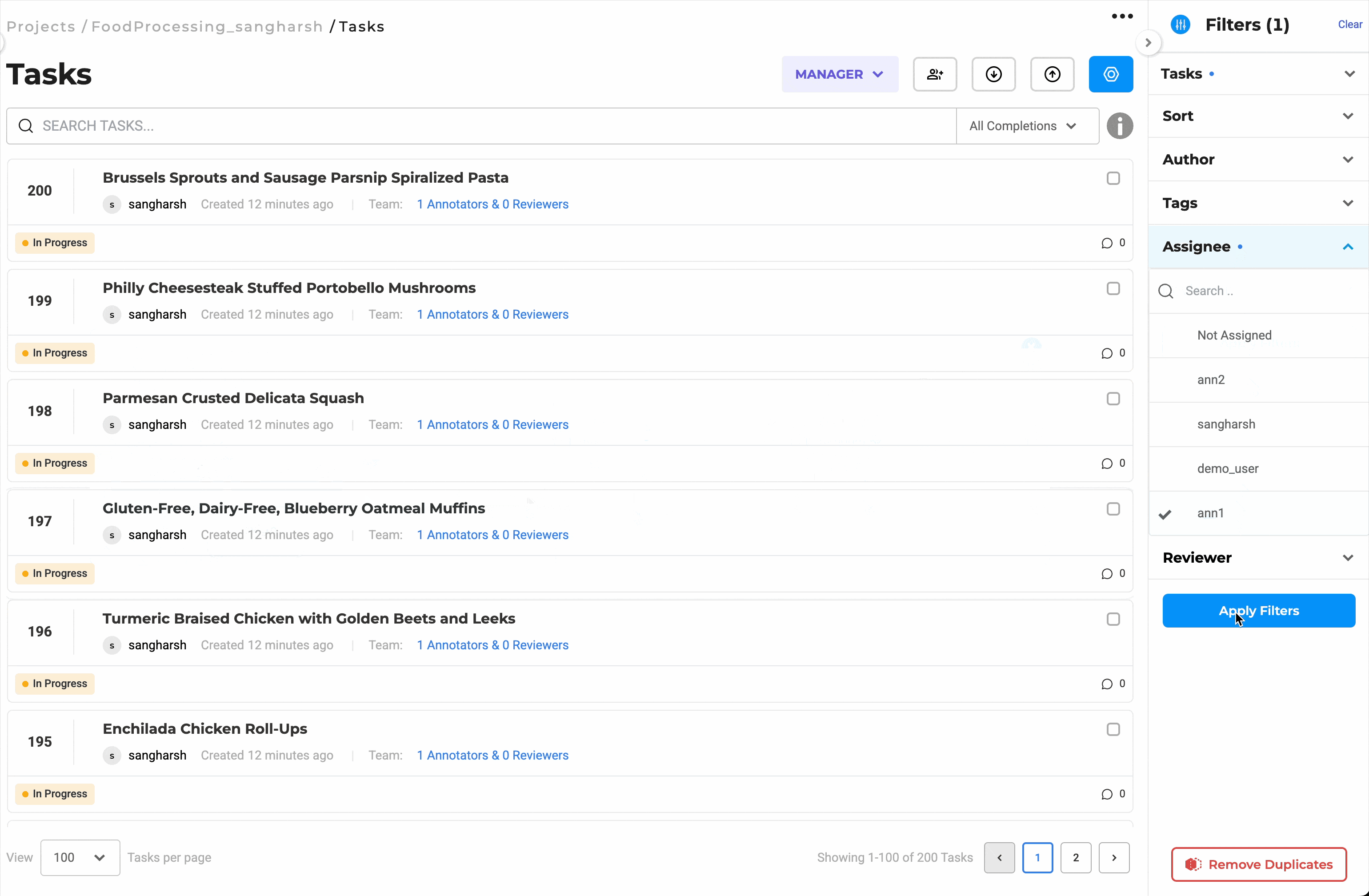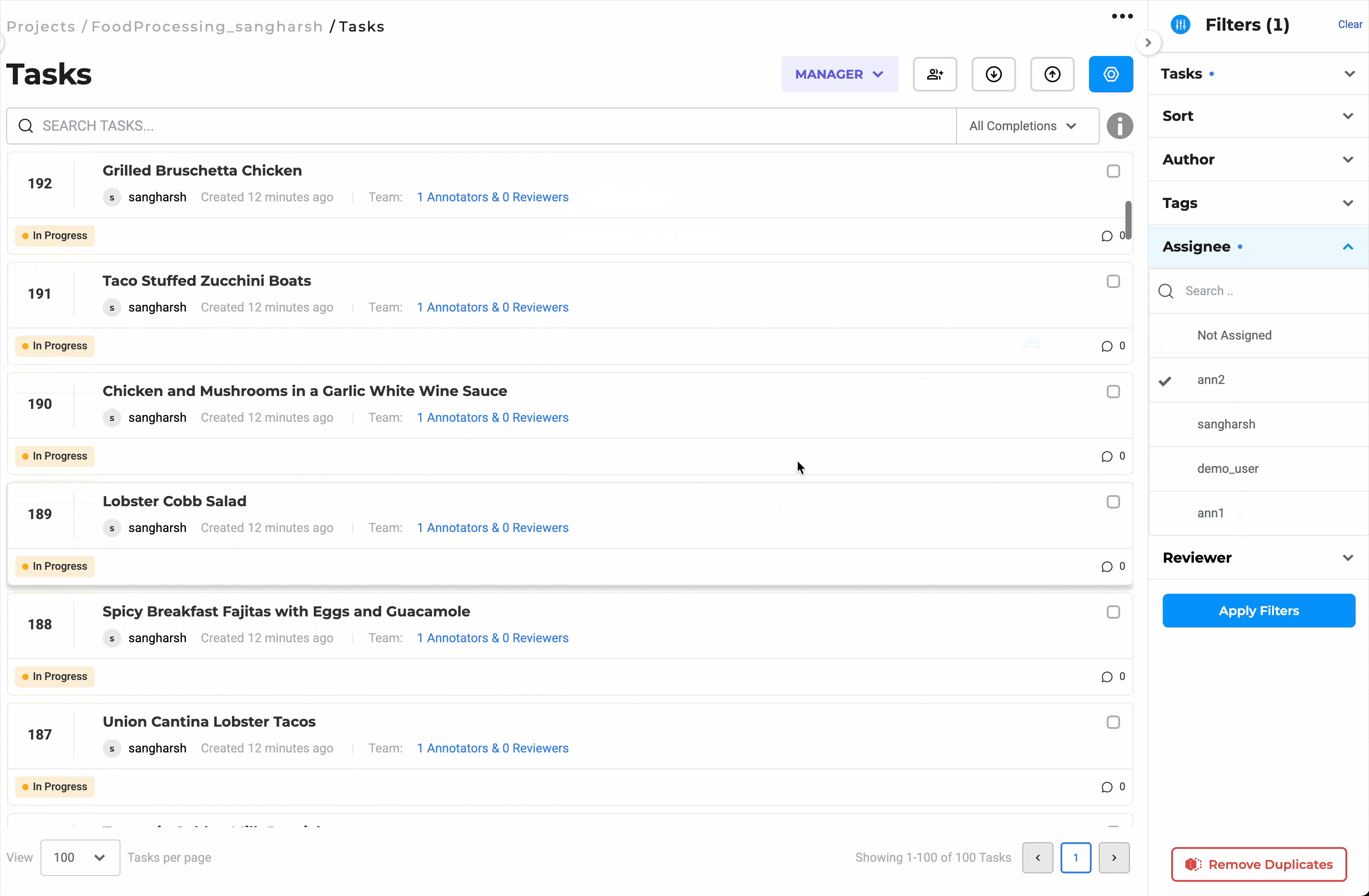
Bulk assignment of up to 200 documents for streamlined review.
Review in an Interface Built for Clinical Context
The platform presents each document alongside its extracted text, helping reviewers evaluate annotations in context. Color-coded labels, persistent zoom levels, and remembered scroll positions make it easier to work accurately over extended review sessions.
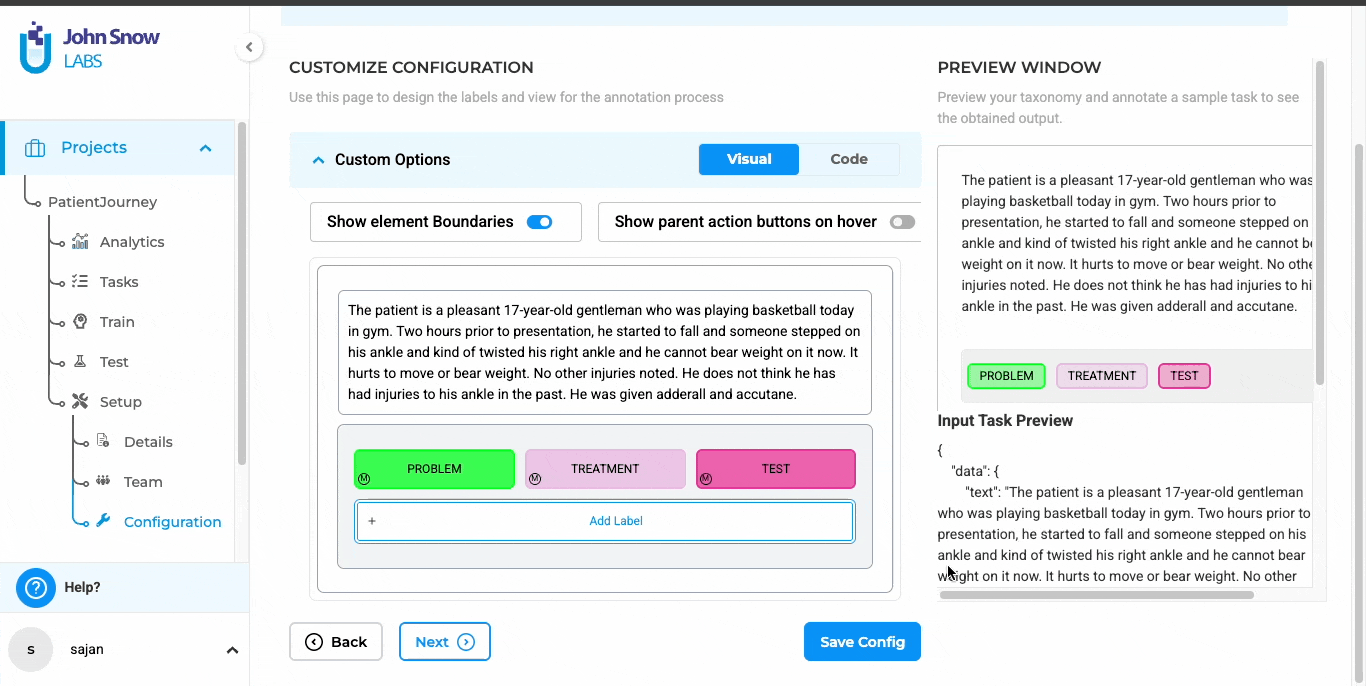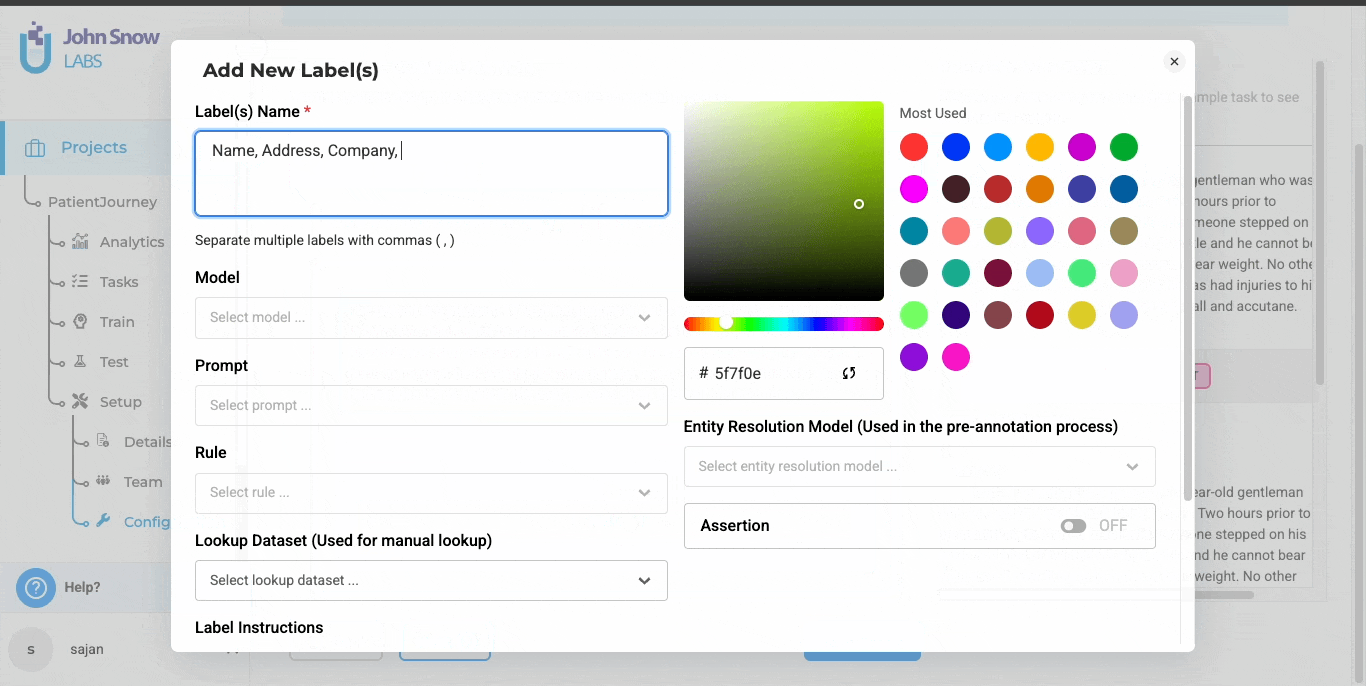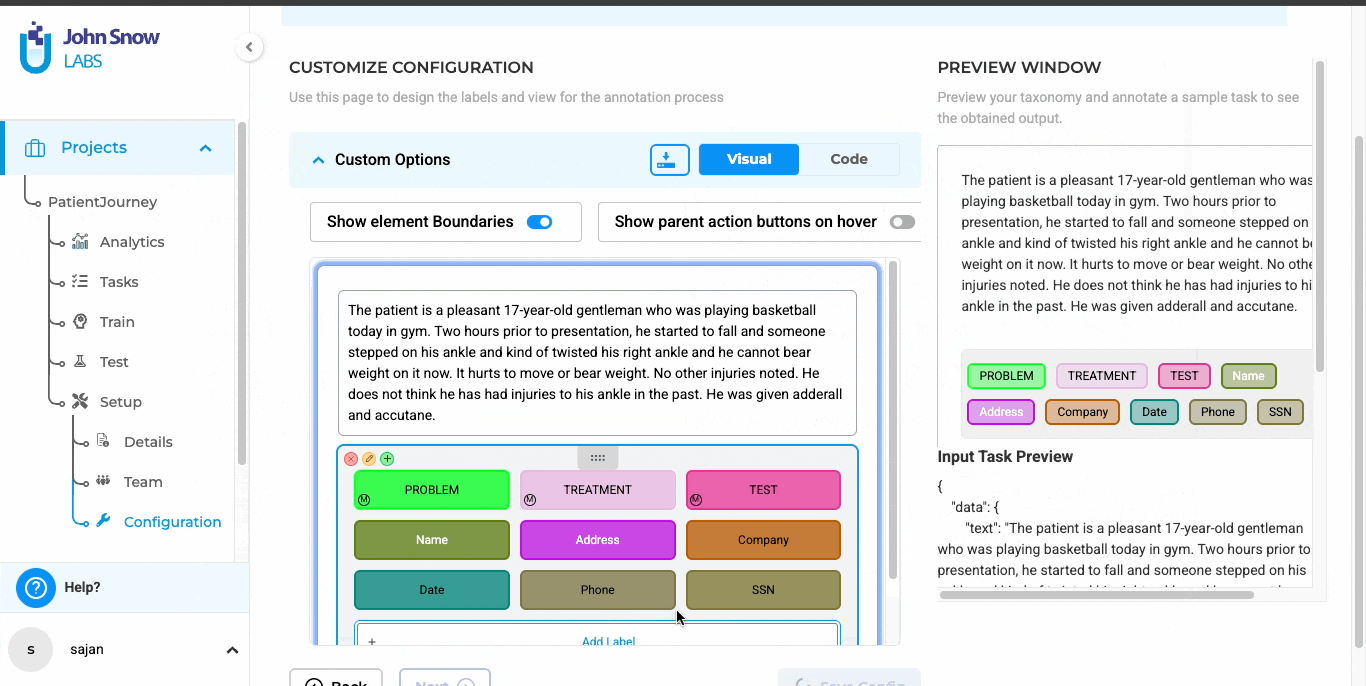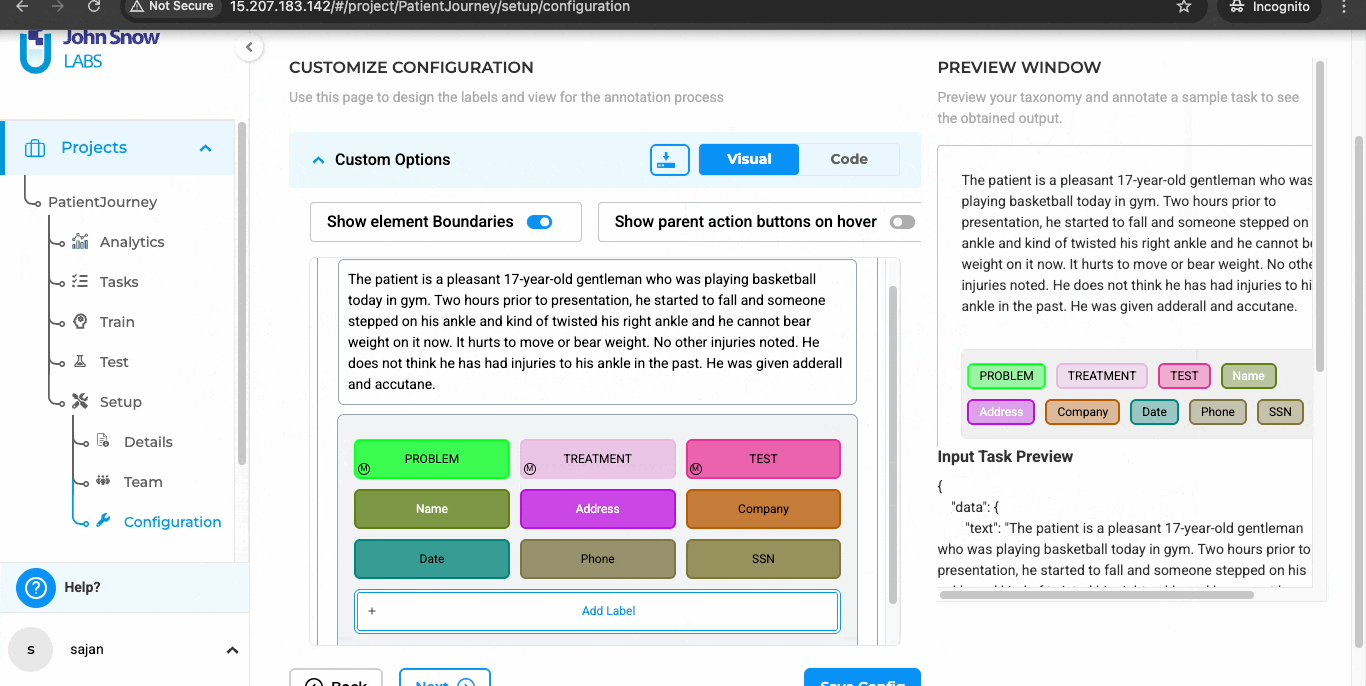
Color-coded labels in Generative AI Lab for faster, more accurate reviews.
Validate and Apply Masking in Two Steps
For text-based de-identification, reviewers confirm whether each tagged entity is actually PHI before applying masking or surrogate replacements. This two-step process reduces the risk of over-masking and provides a clear record of human oversight.
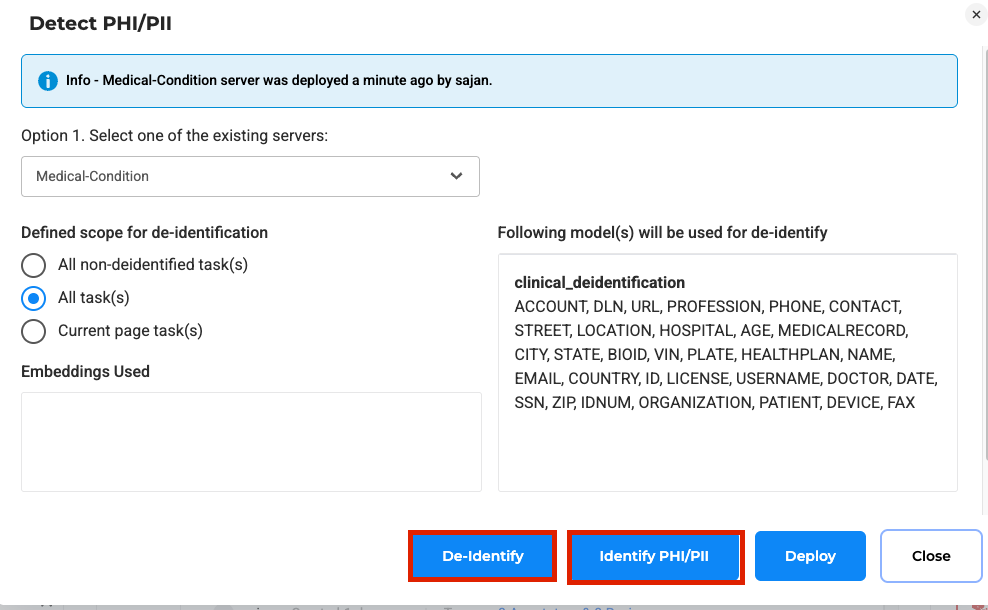
Splitting PHI de-identification into tagging and masking steps.
Export De-Identified Results with a Full Review History
Once the review is complete, you can export a standardized package that includes both the redacted dataset and a full audit log. This export is structured to meet documentation needs for regulators, auditors, and external partners. If you are working with visual data, logs can still be exported, though masking is handled through other tools.
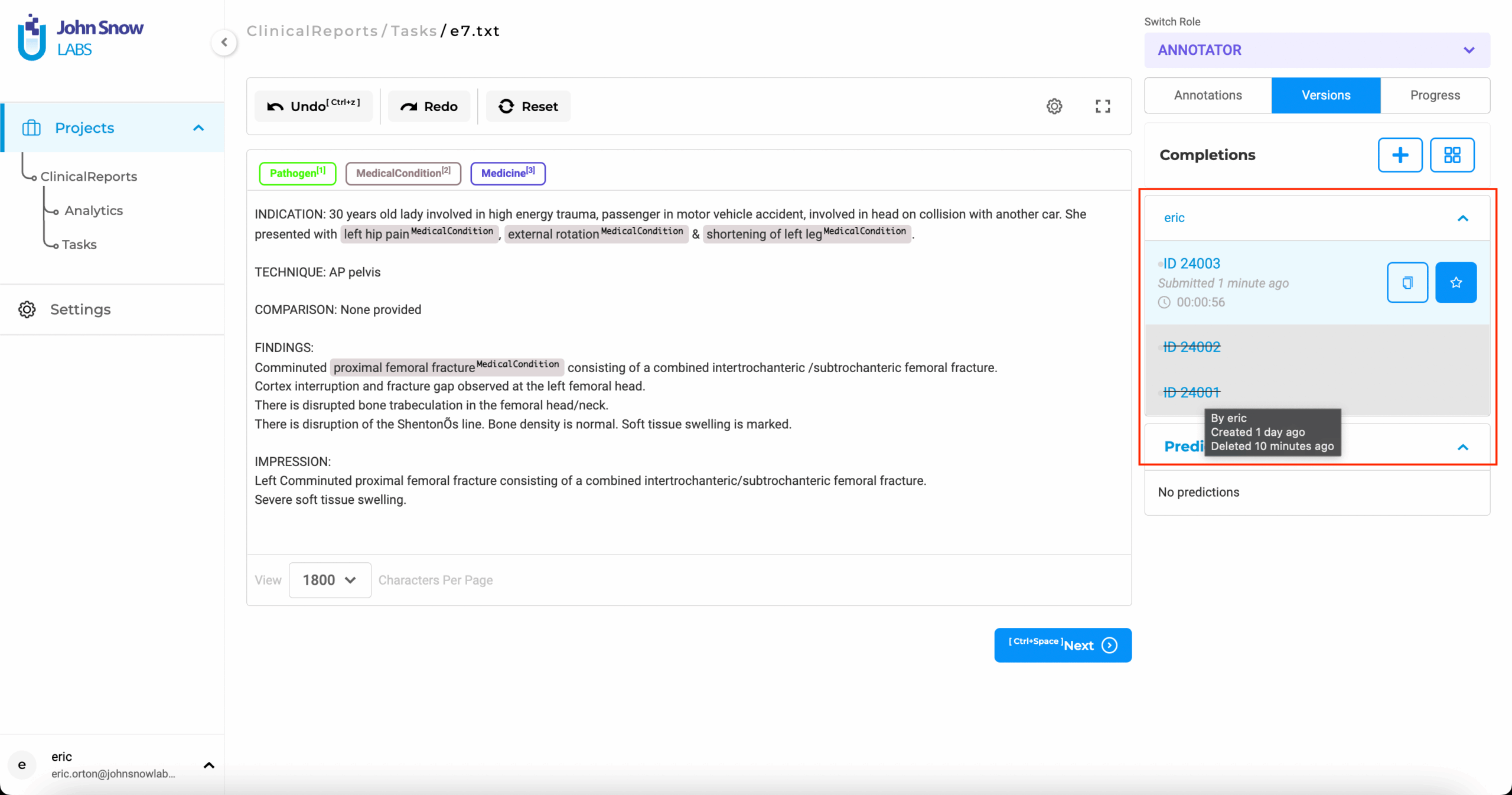
Audit-ready PHI & PII annotation with user-level tracking and strict access control.
Complete PHI Review with an Authorized Validation Team
PHI-cleared reviewers, such as compliance officers or clinical coders, complete the final validation. Every reviewer action is automatically logged to support regulatory requirements and internal audit needs. This step ensures that only validated, policy-compliant data is approved for release.
Reuse Privacy-Safe Data in Follow-Up Projects
After PHI is removed and validation is complete, the dataset can be safely used in new projects. These can include NER annotation, model training, or other analytics workflows. Because the data no longer contains PHI, it can be accessed by broader teams without additional compliance concerns.
Deploy the Platform in Your Own Infrastructure
Generative AI Lab can be installed in your environment using a Helm chart. It supports deployment on AWS, Azure, private Kubernetes clusters, or even fully air-gapped systems. All model inference and PHI processing stay inside your infrastructure, supporting your data security and residency policies.
Reset Projects Quickly Without Losing Your Setup
When requirements change or new data arrives, use the bulk-reset option to clear existing tasks while preserving project settings. This allows your team to restart work immediately without needing to reconfigure the project from scratch.
Why Generative AI Lab Stands Out
Generative AI Lab offers three core capabilities that help healthcare organizations move faster, stay compliant, and scale with confidence.
Enterprise Scalability
Many healthcare teams face hard limits when trying to process large volumes of data using public cloud services. Generative AI Lab sidesteps those ceilings by running Spark‑native pipelines inside your environment. Independent benchmarks show the platform keeps licensing costs steady even when throughput rises to a billion clinical notes.
Public‑cloud tools, by contrast, handle one document at a time in synchronous mode and switch to slower batch jobs for larger volumes. With Generative AI Lab, hospital‑scale workloads clear overnight on infrastructure you already control, and protected data never leaves approved boundaries.
Context‑Aware Automation
Generative AI Lab enhances the Reuse Resource interface by displaying only compatible models, prompts, and rules based on the selected project type. This reduces configuration errors and eliminates unnecessary trial and error.
Teams can start projects faster, reduce rework, and maintain cleaner audit trails. As new use cases are added, this built-in guidance helps maintain consistency across teams and projects.
Integration‑Ready APIs
Every action available in the user interface is also accessible through a fully documented REST API. IT teams can connect Generative AI Lab directly to existing data pipelines, governance dashboards, and compliance reporting systems without building additional tools.
A single surface for both UI and code accelerates compliance checks, simplifies maintenance, and keeps data governance visible throughout the project lifecycle.
FAQs
What infrastructure is required to run the Generative AI Lab on-premises?
The Generative AI Lab is installed using Helm and runs on any Kubernetes cluster, making it easy to deploy within existing infrastructure. The minimum requirements for on-premise use are an 8-core CPU, 32 GB of RAM, and 512 GB of SSD storage.
For teams building or validating text-based models with AI-assisted pre-annotation, the recommended setup is a 16-core CPU, 64 GB of RAM, and a storage footprint of the same size.
For Document Understanding or visual model training workflows, the high-performance configuration includes 4 GPUs, 48 CPU cores, and 192 GB of RAM, equivalent to an AWS g4dn.12xlarge instance.
Can Generative AI Lab remove PHI from medical images and DICOM metadata?
Yes. The Visual NLP module uses OCR to extract PHI from both image pixels and DICOM headers. Once extracted, these elements are processed through the same masking and redaction logic applied to text.
This integrated approach ensures that privacy policies are consistently enforced across all data types while enabling parallel processing of large imaging datasets using Apache Spark.
How well does Generative AI Lab integrate with existing data pipelines and compliance tools?
Every button in the UI maps one-to-one to a REST endpoint, documented under API Integration. That means you can auto-push tasks from an HL7 feed, stream audit logs into Splunk, or trigger model retraining from your CI/CD pipeline with the same calls the front-end makes.