Over the past few years, there’s been a quiet revolution in healthcare AI. The buzz around large language models (LLMs) has pushed them into the limelight, capturing imaginations with their ability to chat fluently, answer complex questions, and summarize lengthy clinical notes.
But when the focus shifts from demos to deployment, a more layered picture begins to emerge. In the context of real-world healthcare where precision isn’t optional, workloads scale into millions of records, and terminology accuracy is non-negotiable different trade-offs come into view.
Some teams have spent years refining tools purpose-built for this environment. Take, for instance, model libraries that prioritize domain specificity, transparency, and integration into existing clinical workflows. With thousands of pretrained components, including dedicated modules for tasks like named entity recognition, these tools aren’t designed to impress in conversation, they’re designed to deliver in real life.
So as benchmarks accumulate and new options proliferate, it’s worth stepping back to ask: what are we really optimizing for in clinical NLP? Fluency, or factual alignment? Flexibility, or traceability? Novelty, or trust? Are LLMs really better?
Sometimes, the right answers begin with better questions.
Clinical Named Entity Recognition: When breadth and depth matter
Identification of clinical entities that are named in a clinical note is a foundational task. Without it, downstream tasks, like cohort identification, pharmacovigilance, or risk stratification, are impossible. General-purpose LLMs like GPT-4.5 and Claude 3.7 Sonnet can identify entities with a decent level of recall, especially if prompted skillfully. But when it comes to extracting nuanced medical concepts as distinguishing between symptoms, diagnoses, and imaging findings, or teasing out dozens of lab test results these models start to fail.
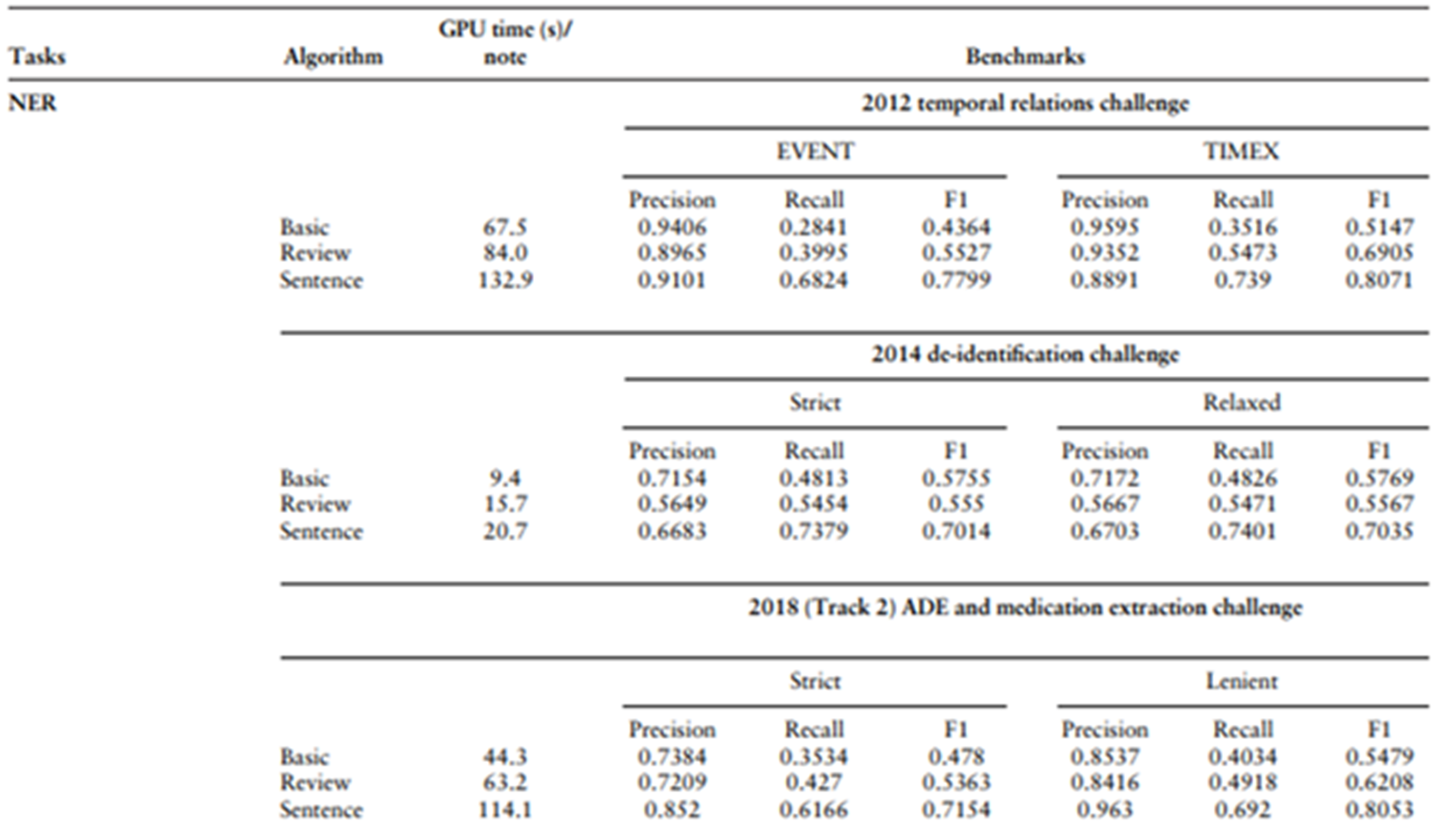
Despite the growing enthusiasm surrounding large language models, their performance in named entity recognition (NER) remains underwhelming when scrutinized closely. Recent benchmarks[1] from the LLM-IE Python package, using Llama-3.1-70B in an 8-shot prompting setting, revealed that the Sentence Frame Extractor only achieved an F1 score of 0.7799 for event entities and 0.8071 for temporal expressions in the i2b2 2012 challenge.
In the 2014 i2b2 de-identification task, strict F1 scores hovered around 0.70. And in the 2018 n2c2 ADE and medication extraction challenge, the strict F1 reached just 0.7154, hardly competitive with classical domain-specific models, which consistently outperform these LLM approaches while demanding far fewer resources. Although the lenient F1 scores nudged above 0.80, such metrics obscure the fundamental limitations: lower recall, susceptibility to prompt variability, and lack of transparency.
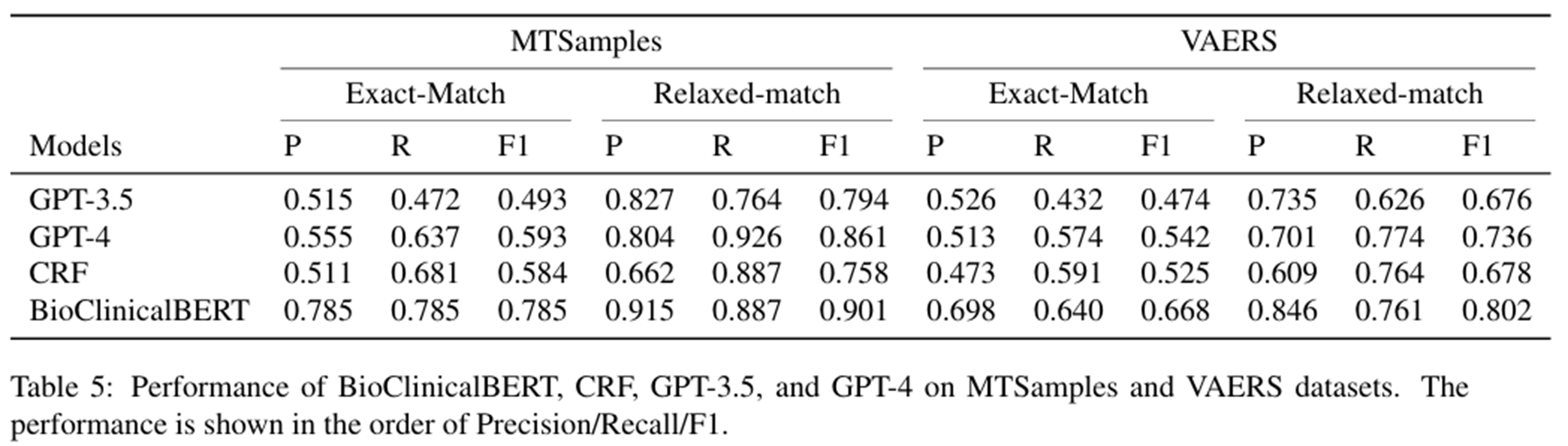
Another study published in September of 2024[2] explored the performance of GPT LLMs as GPT-3.5 and GPT-4 when recognizing clinical entities in unstructured texts after carefully prompting design finding that methods like conditional random fields (CRFs) or Clinical BERT token classifiers outperformed LLMs on this task
All these results reinforce a clear message. While LLMs hold promise and demonstrate impressive versatility in some domains, their current trajectory in core tasks like NER suggests they are not yet ready to displace the robust, fine-tuned pipelines built on traditional biomedical NLP frameworks. Furthermore, while general LLMs are easy to prototype with, their cost, latency, and inconsistency make them impractical on a scale. Extracting three simple facts from 100 million patient notes could cost hundreds of thousands of USD and take several days using GPT-4.5.
John Snow Labs vs General-Purpose Large Language Models
With a more than a decade track of successful real-life deployments, the suite of John Snow Labs healthcare entity recognition models and pipelines covers more than 400 specific entity types through over 130 dedicated models. These solutions offer a high level of detail including fine-grained categories like 20 disorder subtypes.
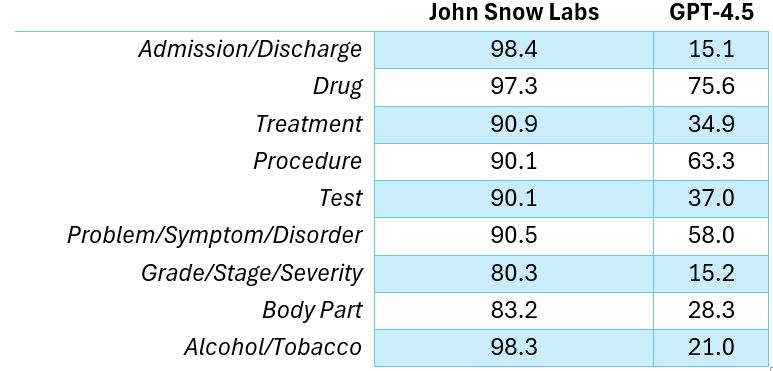
In a benchmark comparing John Snow Labs models against GPT-4.5 on nine common clinical entity types, these models made five to six times fewer errors.
F1 Scores for different entities recognition.
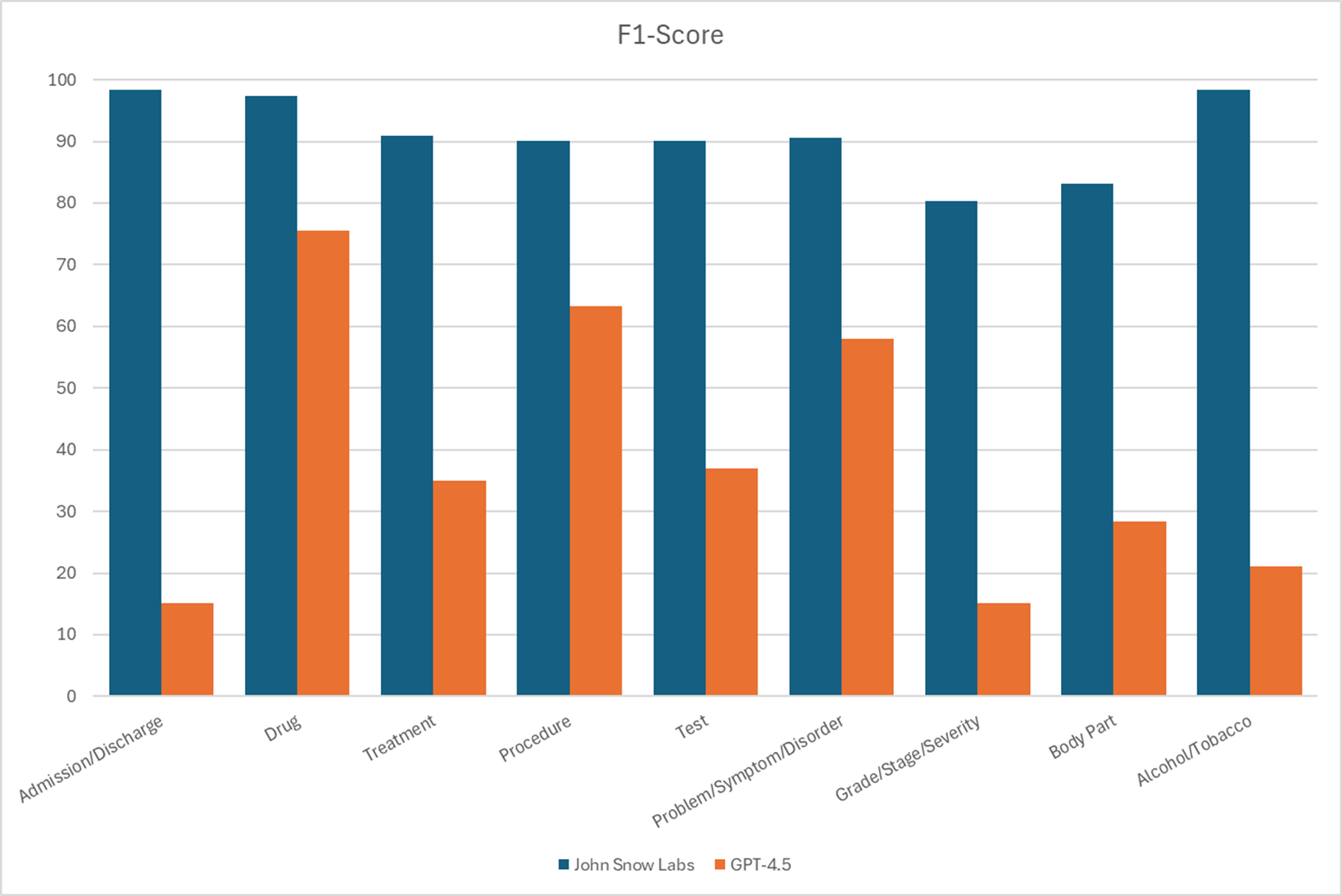
F1 Scores for different entities recognition.
For clinical entity extraction, John Snow Labs’ models make five to six times fewer errors than GPT-4.5.
The same models were compared with Claude 3.7 Sonnet producing five to six times fewer errors than the LLM from Anthropic when extracting the same clinical entities.
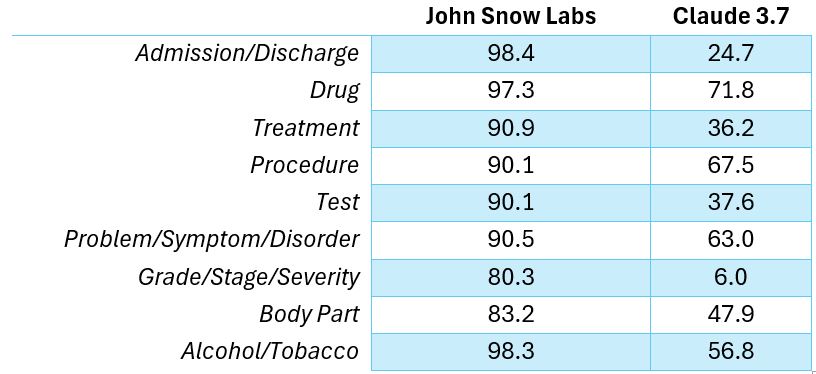
F1 Scores for different entities recognition.
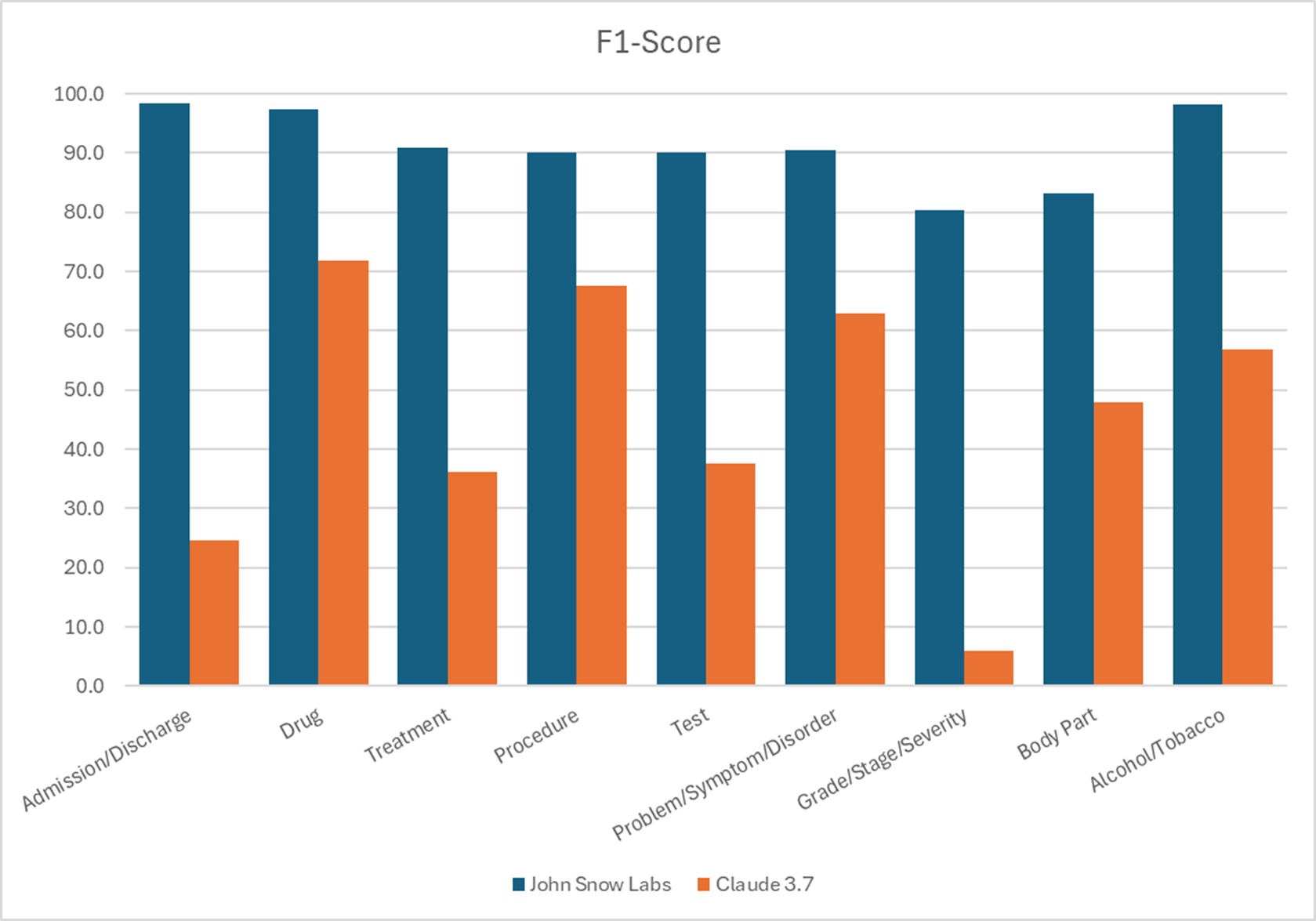
F1 Scores for different entities recognition.
John Snow Labs’ models made five to six times fewer errors than Claude 3.7 Sonnet in clinical entity extraction.
Assertion Status Detection: When “Yes” Doesn’t Mean Yes
Merely identifying that “diabetes” is mentioned in a note isn’t enough. Was it affirmed? Denied? Hypothetical? Historical? Perhaps it referred to the patient’s father. For example, “the patient gets shortness of breath only during exertion” is a conditional statement that influences clinical interpretation. This is where assertion status detection comes in, a subtle but essential task.
While LLMs can guess the intent behind a phrase, their output is inherently non-deterministic and not easily auditable. Clinical domain-specific assertion models, on the other hand, are built on multiple architectural backbones, from Bi-LSTM to transformer classifiers and rule-based logic, and support assertion types like “possible,” “conditional,” and “associated with someone else.” This modularity allows an even more specialized fine-tuning for subdomains like oncology or radiology, where different clinical nuances matter.
John Snow Labs’ models outperform GPT-4.5 in detecting clinical assertion status, with greater accuracy and transparency across medical domains.
John Snow Labs has developed a modular framework for clinical assertion detection that combines large language models, deep learning, few-shot learning, and rule-based methods into a unified, production-ready pipeline within Healthcare NLP library. A fine-tuned Large Language Model adapted using LoRA and trained on the i2b2 dataset, delivers top-tier accuracy across most assertion types but requires substantial GPU resources and has slower inference speeds.
To provide faster alternatives, the framework includes AssertionDL, a Bi-LSTM model optimized for contextual windows around entities, which performs well on less frequent categories like “conditional” and “associated with someone else.” It also incorporates BioBERT-based sequence classifiers for efficient contextual classification and a few-shot model built on the SetFit framework for scenarios with limited labeled data. Additionally, a customizable rule-based system enhances interpretability and precision in clinical settings.
These models are integrated into a pipeline that shares components and merges outputs using voting and performance-based prioritization. The result is a scalable, cost-effective solution that offers strong accuracy and transparency, outperforming LLMs systems in both flexibility and clinical relevance.
In a paper published on March 2025 [3] John Snow Labs solution delivered higher accuracy when compared with GPT-4o in detecting assertion status, while offering greater flexibility and transparency. Furthermore, these models can run at a fraction of the computational cost of LLMs as GPT-4o.
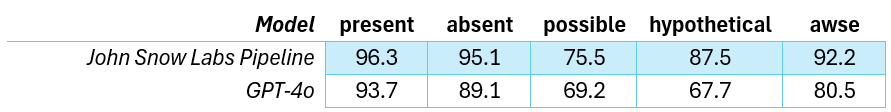
F1-Scores for different assertion categories. awse: associated with someone else.
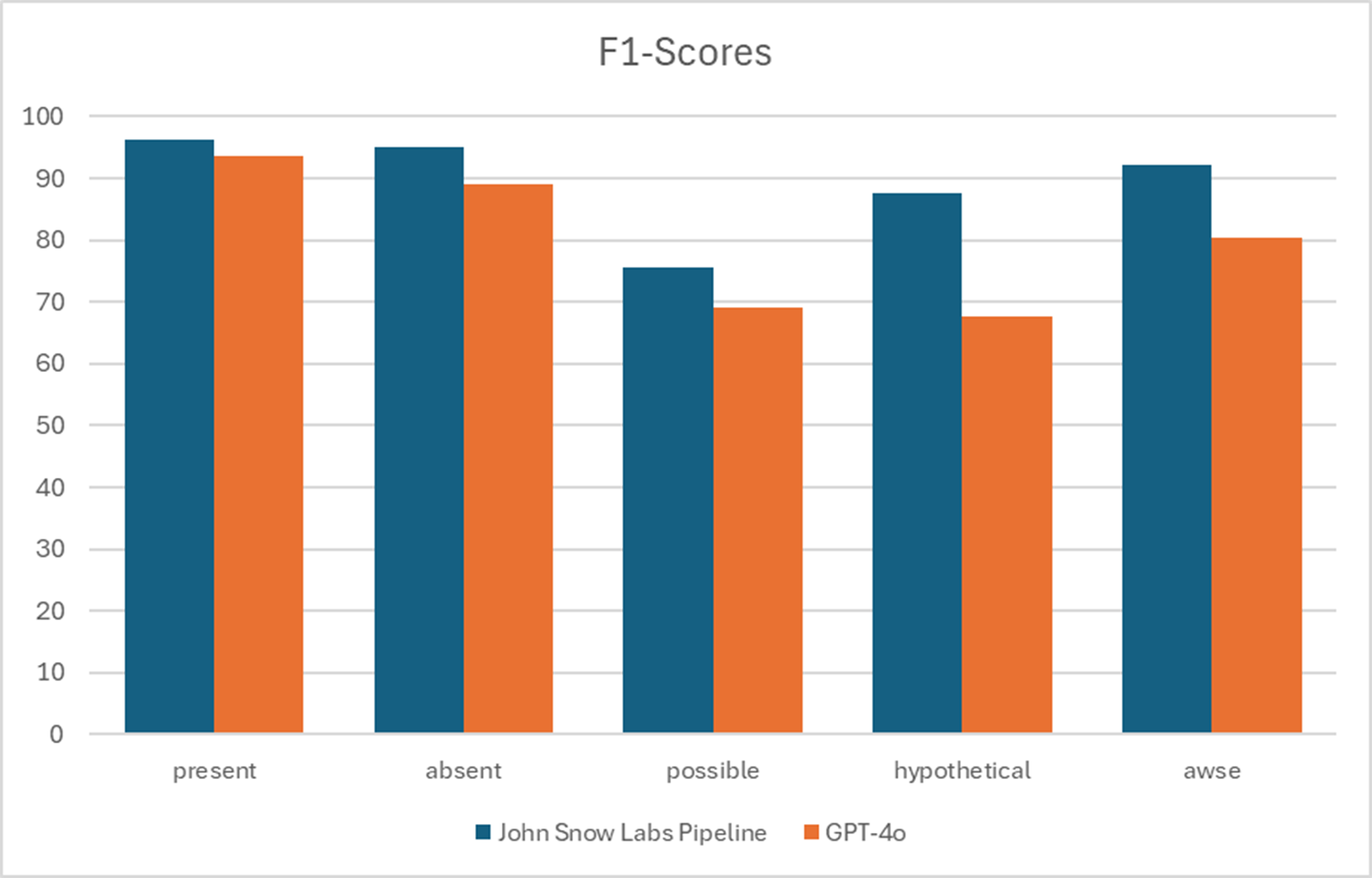
F1 Scores for assertion status. awse: associated with someone else.
Terminology mapping: Not a guessing game
Mapping clinical entities to standard terminologies like ICD-10, SNOMED CT, or RxNorm are not just a technical step, but a critical foundation for downstream tasks like billing, cohort selection, and real-world evidence. What if the cost of a mismatch wasn’t just a formatting error, but a missed diagnosis or misclassified case?
Large language models bring impressive fluency to text generation and summarization, but they aren’t designed to function as deterministic lookup engines. When faced with a terminology mapping task, they may infer rather than resolve, offering plausible guesses instead of precise matches. In contrast, approaches that combine symbolic reasoning, domain-tuned embeddings, and knowledge graphs may offer a better fit.
For instance, a recent RxNorm benchmark showed that the vector-based terminology resolver from John Snow Labs, which leverages domain-specific models, achieved 85% top-five accuracy. GPT-4o, by comparison, reached just 9%. And while the customized approach would process one million records for about $4,500, the LLM-based alternative would cost more than $22,000, while delivering significantly lower accuracy.
That is why, in tasks where precision is critical, it’s worth asking: is generative AI the best fit or would a purpose-built approach be more reliable?
John Snow Labs’ terminology mapping solution achieved 85% accuracy, nearly ten times higher than GPT-4o, at just one-fifth the cost.
Tools built for purpose win at scale
There’s much to appreciate about large language models. Their fluency, adaptability, and breadth make them compelling for tasks like creative summarization and open-domain Q&A. But when the task is specific, the scale is industrial, and the domain is clinical, general-purpose models often fall short.
However, multiple benchmarks consistently show that domain-adapted, well-engineered pipelines, especially those designed for clinical language, outperform generic LLMs in accuracy, transparency, and cost-efficiency. They are faster, more interpretable, and more cost-effective at scale.
So before reaching for the latest general-purpose LLM API, ask: is this a space for experimentation or one that demands reliability? In high-stakes environments like healthcare information extraction, the answer often favors proven, purpose-built tools.
References
[1] Hsu E, Roberts K. LLM-IE: a python package for biomedical generative information extraction with large language models. JAMIA Open. 2025 Mar 12;8(2):ooaf012. https://pmc.ncbi.nlm.nih.gov/articles/PMC11901043/
[2] Hu Y, Chen Q, Du J, Peng X, Keloth VK, Zuo X, Zhou Y, Li Z, Jiang X, Lu Z, Roberts K, Xu H. Improving large language models for clinical named entity recognition via prompt engineering. J Am Med Inform Assoc. 2024 Sep 1;31(9):1812-1820. https://pubmed.ncbi.nlm.nih.gov/38281112/
[3] Kocaman, V., Gul, Y., Kaya, M. A., Haq, H. U., Butgul, M., Celik, C., & Talby, D. (2025). Beyond Negation Detection: Comprehensive Assertion Detection Models for Clinical NLP. https://arxiv.org/abs/2503.17425