Clara Ferrer still remembers the morning the call came from the regional cancer registry. The researchers had finally received money for a real world study on new cancer drugs, and they wanted her international hospitals group to share ten years of electronic notes, x ray headers and lab PDFs. Clara, the hospital’s head of data governance, felt both pride and worry. Pride, because her hospital group’s records could help thousands of future patients. Worry, because she had been through this before: millions of files, many note styles, and the fear that one missed name or phone number could leak and break the trust of patients and regulators.
That tension is familiar to anyone who guides electronic health records into research projects. You stand between eager scientists and the private stories your doctors write each day. You know the rule books from HIPAA to the GDPR, and you have chaired more de identification meetings than you can recall. Yet the basic problem still remains. A medium sized health system can collect a terabyte of free text every year. Manually hiding personal facts at that scale is slow and costly, but a black box computer tool feels risky when you must sign the final compliance form.
What the big LLM vendors promise
Over the past year several tech giants have offered a silver bullet story. Their plan: use one large language model and a retrieval step that adds the HIPAA rules and a few regular expressions. Press the button, they say, and the model will read every note, mark any private detail, and return a clean copy. It sounds great, especially if the same system already runs your patient chatbot.
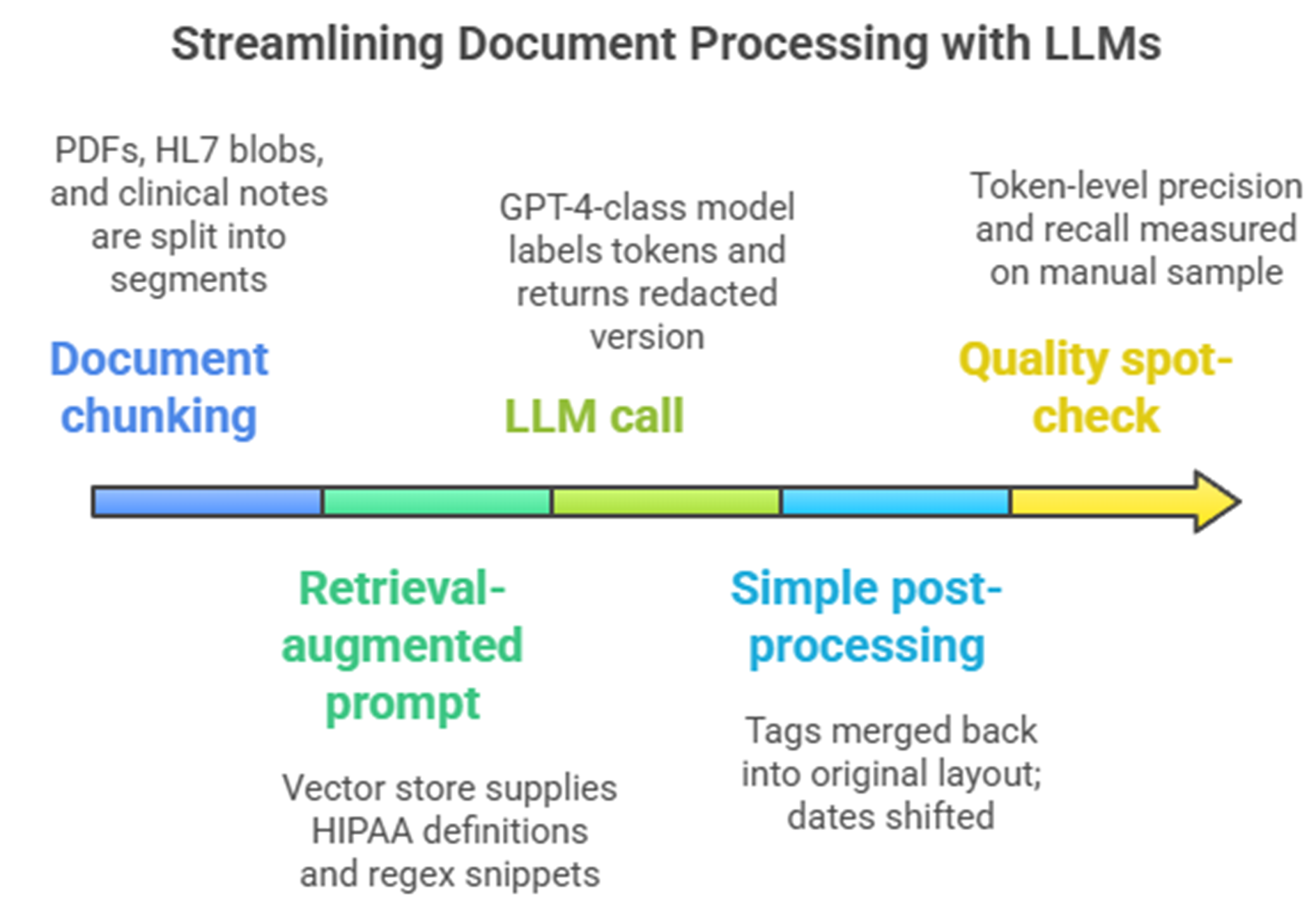
Typical single GPT + RAG de-identification setup
Look closer and the shine fades. Independent tests show that even GPT‑4, with careful prompts, finds only about 83% of private details in real clinical notes, leaving almost one in five exposed. A head‑to‑head test on forty‑eight multi‑site documents saw a zero‑shot GPT‑4o pipeline reach an F1 of 79%, while Azure’s health API climbed to 91% [1]. Both numbers miss the expert‑approval bar and cause cloud costs that top 500,000 USD [1] for an 8 million notes job. These general models also stumble on many languages and note styles. Dates in Spanish letters slip through, French ID numbers confuse the parser, and the made‑up names vary from note to note, breaking long‑term links for research. Because these models sample by design, running the same file twice can give different results, making audits hard.
Limitations of single GPT + RAG de-identification setup
| Aspect | Most likely outcome with a single GPT + RAG setup | Proof behind the view |
| PHI recall | About 80–90 % for clear IDs; lower on people names, building names or foreign addresses | GPT‑4 recall 0.83 on real notes with strong prompts [2] |
| False positives | Fair amount; drug names like “Atenolol” or places like “Jordan” often hidden, hurting later NLP | Generic models lack medical vocabulary; radiology study showed mixed results yet still missed names [3] |
| Surrogate consistency | Not steady across files; “Dr Smith” may become “Dr Brown” in one note and “Dr Lee” in another, breaking long‑term links | Retrieval step cannot hold memory across notes; special pipelines use tables for stable surrogates [4] |
| Cost profile | $0.001–0.002 per 1 000 tokens – about $500–$800 K for eight million notes (≈ 1.2 TB) | Azure and GPT‑4o quotes showed >$600 K for 1 M short notes; local model cut cost 80 % [1] |
| Speed | Tens of thousands of notes per GPU‑hour; speed scales with GPUs but cloud bills rise with it | No built‑in distributed run; API caps batch size at large scale |
| Auditability / QC | Black‑box logic; hard to get token‑level confidence or repeat runs [beyond-accuracy-automated-de-identification-of-large-real-world-clinical-text-datasets.pdf] | ChatGPT‑like systems 33 % less accurate than domain models and show no probability [4] |
| Language & template drift | Decent on modern English; poor on Spanish, French, German or old ASCII notes | General models trained on web text under‑represent medical formats; recall drops on radiology headers |
| Left‑over privacy risk | Even at 90 % recall, attacks can show if a patient is in the dataset | De‑identified MIMIC notes showed attacker edge 0.47 [5] |
Picture Clara approving a pilot on one month of ENT notes. At first things look fine, until a resident finds a letter that still holds the father’s phone, written as “+34 600 123 456” in Spanish. The model had a token limit and cut the phone patterns for non‑US formats. Another line turns “Dr María Luque” into “Dr Mark Luke,” a made‑up name that fits English but not Spanish, hurting links to earlier visits. Clara’s team is not sure these slips are rare. They would need to review hundreds of files in every language to trust the tool again, adding weeks of work and leaving the doubt that one hidden detail may still sit in the 8 million notes backlog.
The healthcare native John Snow Labs solution
Many studies and real hospital projects point to a different way. Teams at Penn Medicine [3] and several hospital groups [4] show that hybrid pipelines—medical deep‑learning models plus hand rules—beat single LLM setups on both recall and audit ease. In the 2014 i2b2 contest, systems that mixed Conditional Random Fields with special dictionaries passed the 95 % recall bar that regulators like [4]. More recently, the Emory HIDE tool and the NeuroNER network tested on 2 503 radiology reports hit F1 scores over 92 %, well ahead of rule‑only tools and still easy to check [3].
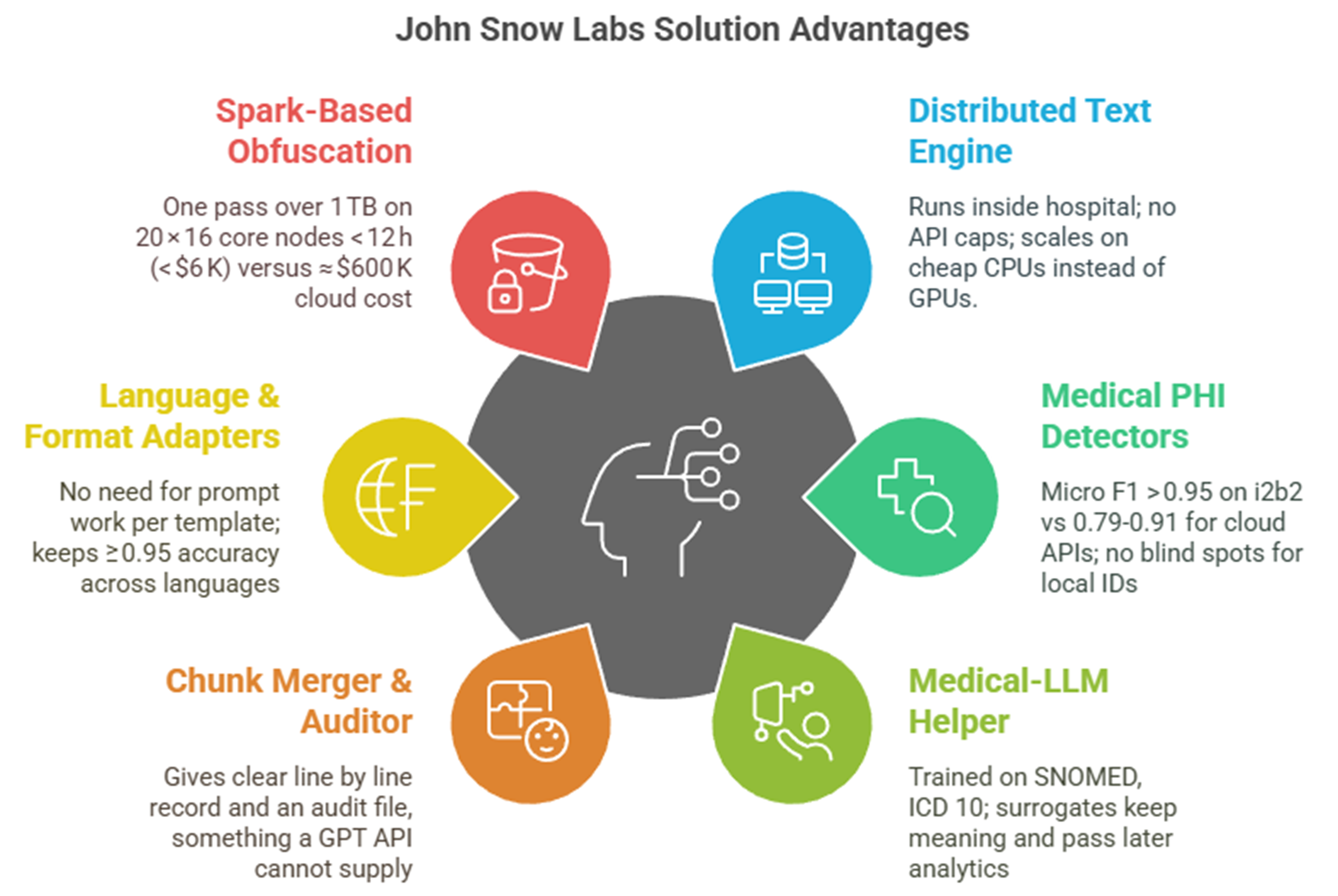
This growing proof suggests that domain‑tuned models with rule guards are not just a sales pitch but good practice. What if such a toolkit were already out there? Several hospitals now use the open Healthcare NLP parts from John Snow Labs, building Spark jobs that blend medical deep‑learning finders with rule‑based safety checks. Early reports say the models spot more than eighty ID types, from ward phone lines to implant medical device numbers, while staying above ninety‑five per cent recall on the i2b2 set and in billion‑note jobs [4]. English, Spanish, French and German models come ready to run, so the same pipeline can scan mixed‑language archives without prompt tricks. When the de‑identifier meets a name or date it cannot simply mask, a Medical‑LLM trained on biomedical text makes a new value that stays the same across all follow‑up visits, which keeps research datasets linked.
Comparison versus John Snow Labs solution and generic GPT + RAG
| Dimension | Healthcare‑aware stack | Generic GPT + RAG |
| PHI recall | 96–98 % token‑level (i2b2 & real‑world use) [4] | 80–90 % – GPT‑4 recall 0.83 on real notes; GPT‑4o F1 0.79 on multi‑site set |
| Cost for 8 M notes (≈ 1.2 TB) | ≈ $90 K one‑off license + $6 K compute run | $500–$800 K in usage charges (GPT‑4/o or Azure) |
| Multilingual ready | Seven languages ready; same pipeline, no prompt edit [4] | Accuracy drops on non‑English notes; prompts need editing |
| Audit & repeat | Fully repeatable Spark job; per‑token confidence + hash chain | Temperature and API updates mean non‑repeatable results, no confidence scores |
| Data value kept | Surrogates from SNOMED/RxNorm and LLM tables; keeps references over time | Surrogates differ per chunk; timing cues can vanish, hurting later studies |
On the tech side, users tell a simple story: the largest JSL Medical LLM with 14b parameter model loads in half precision on a 40Gb GPU , while the rest of the pipeline runs on normal CPUs. Cara Ferrer Hospital’s pilot, for example, could clean 1.2 terabytes overnight on twenty standard servers for under 6,000 USD in compute. Each token leaves with a confidence score, and a reversible hash lets ethics boards unmask if truly needed. In clinical tests the same Medical‑LLMs beat GPT‑4, scoring 89% versus 86% on the OpenMed quiz, a small but calming gap that lowers the risk of wrong drug advice.
A closer look to the John Snow Labs De-Identification solution
Healthcare NLP is not one box but a set of pieces that click together in Spark. Clara’s team starts with DocumentAssembler and the multilingual SentenceDetectorDLModel to keep German umlauts and French accents intact. They then load clinical BERT embeddings (embeddings_clinical_bert) that feed two NER engines: ner_jsl_enriched for 87 clinical labels and ner_deid_generic for ID labels. A NerConverter and ChunkMerger tidy overlaps.
Dates shift safely thanks to DateNormalizer, which parses “14 avril 2024” as easily as “14 April 2024”. For plain hiding, the DeIdentification node masks with letters or stars, but for research the team turns on the Faker mode: this inserts locale‑aware fake values and keeps PDF layout. A small RuleSynthesizer lists extra regexes for Spain’s SIP card and France’s NIR number, all used by ContextualParser, no code changes needed.
To keep the option to re‑identify under ethics review, the last step adds DocumentHashCoder. It salts each note with a SHA‑256 hash of the MRN, stores the key pair in a locked Parquet file and gives analysts only the hash. If the cancer board later needs to know the real MRN, a one‑line ReIdentification job recreates the source text.
Performance is solid. On a single A100 the Medical‑LLM 14B makes surrogates at 900 tokens per second. The rest of the Spark job fills five vCPUs per worker, handling 45,000 notes a minute on the hospital cluster. Using the GPU‑free path raises speed to 60,000 notes a minute but loses the more advanced surrogates.
All parts come under one Healthcare NLP license; no extra fees for languages or the medical LLM. Demo notebooks sit in the public John Snow Labs Workshop public repo so Clara’s data team built a full test job before the lawyers even saw a contract.
The choice for data‑governance teams is not between slow manual work and a single big AI. It is between a one‑size model that almost works and a healthcare‑ready platform already proven at scale. Hospitals using the John Snow Labs approach see fewer misses, lower review time and, most important, more confidence when they share years of patient stories with outside teams. Clara Ferrer, two months after go‑live, now gets weekly progress dashboards instead of late‑night worry calls. Her notes move safely, her budget stays on track and her sleep, at last, is undisturbed.
Generative AI Lab: De-identification with human expert review healthcare
For non-data scientists, the out-of-the-box de-identification capabilities in John Snow Labs’ Generative AI Lab on AWS Marketplace is the way to go. It not only makes the review and approval (human in the loop) process easier, the feedback can also allow subject matter experts to improve the accuracy as the models learn.
The workflow for de-identification using Generative AI Lab begins with the secure storage of clinical data, such as patient notes, lab reports, and medical images in customers’ own AWS environment. John Snow Labs NLP models then analyze these unstructured documents to identify PHI information. This identified PHI information can then be configured to perform masking or obfuscation. While Generative AI Lab handles much of the automation, external systems like cloud storage, EHRs, and patient demographic databases are essential for data input ensuring seamless integration and compliance with healthcare regulations.
[1] V. Kocaman, M. Santas, Y. Gul, M. Butgul, and D. Talby, “Can Zero-Shot Commercial APIs Deliver Regulatory-Grade Clinical Text DeIdentification?,” Mar. 31, 2025, arXiv: arXiv:2503.20794. doi: 10.48550/arXiv.2503.20794.
[2] B. Altalla’ et al., “Evaluating GPT models for clinical note de-identification,” Sci Rep, vol. 15, no. 1, p. 3852, Jan. 2025, doi: 10.1038/s41598-025-86890-3.
[3] J. M. Steinkamp, T. Pomeranz, J. Adleberg, C. E. Kahn, and T. S. Cook, “Evaluation of Automated Public De-Identification Tools on a Corpus of Radiology Reports,” Radiology: Artificial Intelligence, vol. 2, no. 6, p. e190137, Nov. 2020, doi: 10.1148/ryai.2020190137.
[4] V. Kocaman, H. U. Haq, and D. Talby, “Beyond Accuracy: Automated De-Identification of Large Real-World Clinical Text Datasets,” 2023, doi: 10.48550/ARXIV.2312.08495.
[5] A. R. Sarkar, Y.-S. Chuang, N. Mohammed, and X. Jiang, “De-identification is not enough: a comparison between de-identified and synthetic clinical notes,” Sci Rep, vol. 14, no. 1, p. 29669, Nov. 2024, doi: 10.1038/s41598-024-81170-y.