Spark NLP for Healthcare De-Identification module demonstrates superior performance with a 93% accuracy rate compared to ChatGPT’s 60% accuracy on detecting PHI entities in clinical notes.
Organizations handling documents containing Protected Health Information (PHI) are obliged to comply with privacy regulations. De-identification of PHI plays a crucial role in enabling the sharing of health data for various purposes, such as medical research, policy assessments, and other evaluations, without infringing on patient privacy or necessitating individual authorizations. Once data is properly de-identified, restrictions imposed by HIPAA and GDPR no longer apply. Common identifiers within PHI include names, locations, dates, phone numbers, email addresses, ID numbers, Social Security numbers, and medical record numbers, among others.
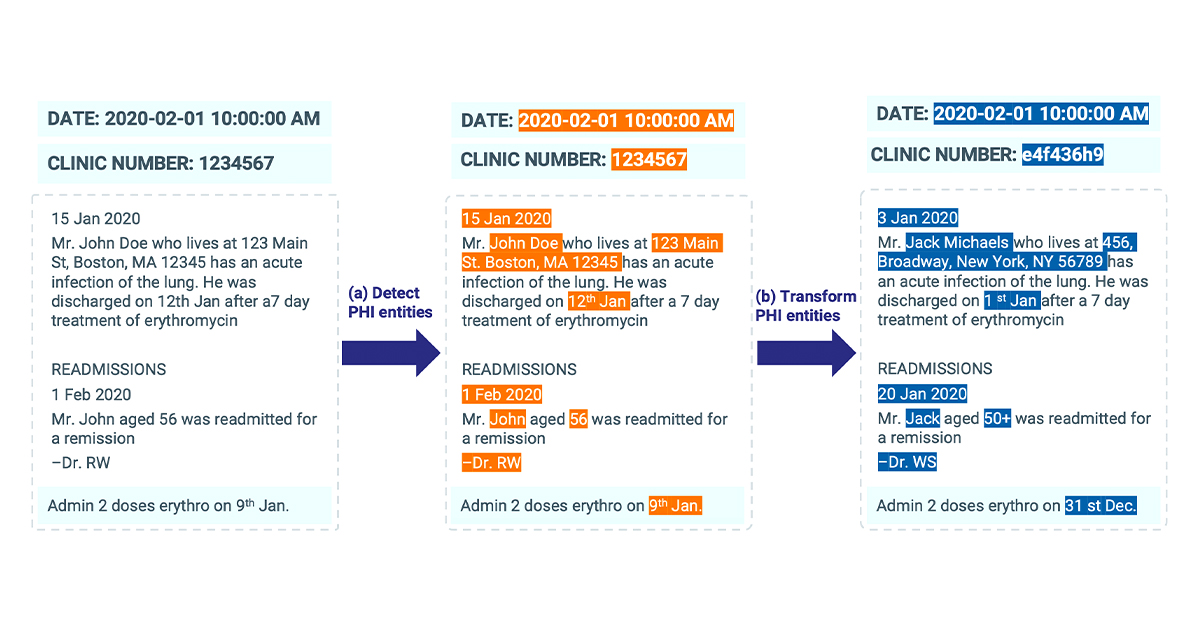
De-Identification process identifies potential pieces of content with personal information about patients and remove them by replacing with semantic tags or fake entities
Using LLMs for de-identification of sensitive data (PHI) could be considered both an overkill and potentially unreliable, depending on the specific use case and the level of customization needed. LLMs, like ChatGPT, are powerful tools that can generate high-quality text but may not be specifically optimized for the task of de-identifying PHI.
De-identification requires precise detection and removal or anonymization of sensitive information. Relying on an LLM to perform this task could result in occasional inaccuracies or missed instances of sensitive data. On the other hand, dedicated de-identification tools or models, such as Spark NLP for Healthcare, are specifically designed for this purpose and are more reliable.
Despite the concerns, some users are still interested in using Healthcare LLMs for de-identification tasks due to the simplicity of the process (e.g., providing a prompt and text, then receiving a de-identified version). As such, we aimed to assess ChatGPT’s performance in the de-identification task and compare it to Spark NLP for Healthcare.
Moreover, obfuscation techniques such as length consistency, name consistency, gender consistency, age consistency, clinical consistency, day shift consistency, and date format consistency play a crucial role in maintaining data consistency and usefulness after de-identification, which we will not delve into in this blog post.
De-Identification Capabilities of Spark NLP for Healthcare
The de-identification module Spark NLP for Healthcare involves a sequence of five stages, including text pre-processing and feature generation, named entity recognition, contextual rules, chunk merging, and obfuscation.
Text pre-processing involves steps like document assembling, sentence detection, tokenization, and word embedding generation. The input text is prepared for named entity recognition (NER) to identify PHI tokens and obfuscate PHI chunks. NER models are at the core of de-identification pipelines and are based on a BLSTM architecture.
The contextual rule engine provides flexibility and complements NER models in cases where specific PHI identifier types are not supported. It includes regex matching, prefix and suffix matching, and proximity and length of context matching. The chunk merger merges outputs from models and rules to resolve conflicting or overlapping detections, increasing the pipeline’s overall accuracy.
The final step involves masking or obfuscating the text, preserving the text layout for various file types. The re-identification vault stores the mappings between obfuscated/masked chunks and their original form in an auxiliary column, allowing for future re-identification if necessary.
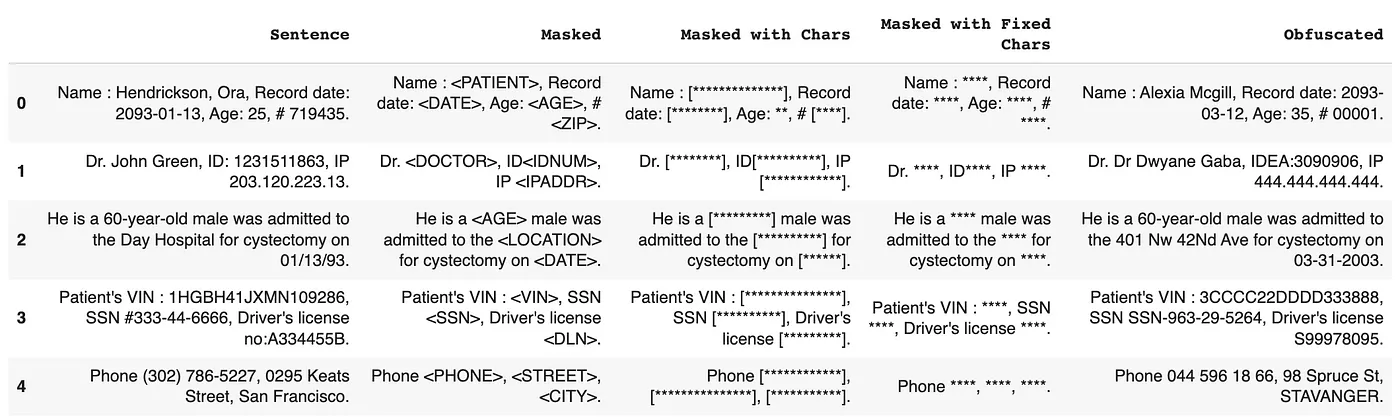
Spark NLP for Healthcare has various options while de-identifying a PHI data
A fully automated solution requires a very high level of accuracy that does not require manual review. A hybrid context-based model architecture is described, which outperforms a single neural network model by 10% on the i2b2–2014 benchmark. The proposed system makes 50%, 475%, and 575% fewer errors than the comparable AWS, Azure, and GCP services respectively. It exceeds 98% coverage of sensitive data across 8 European languages, without a need for fine tuning.
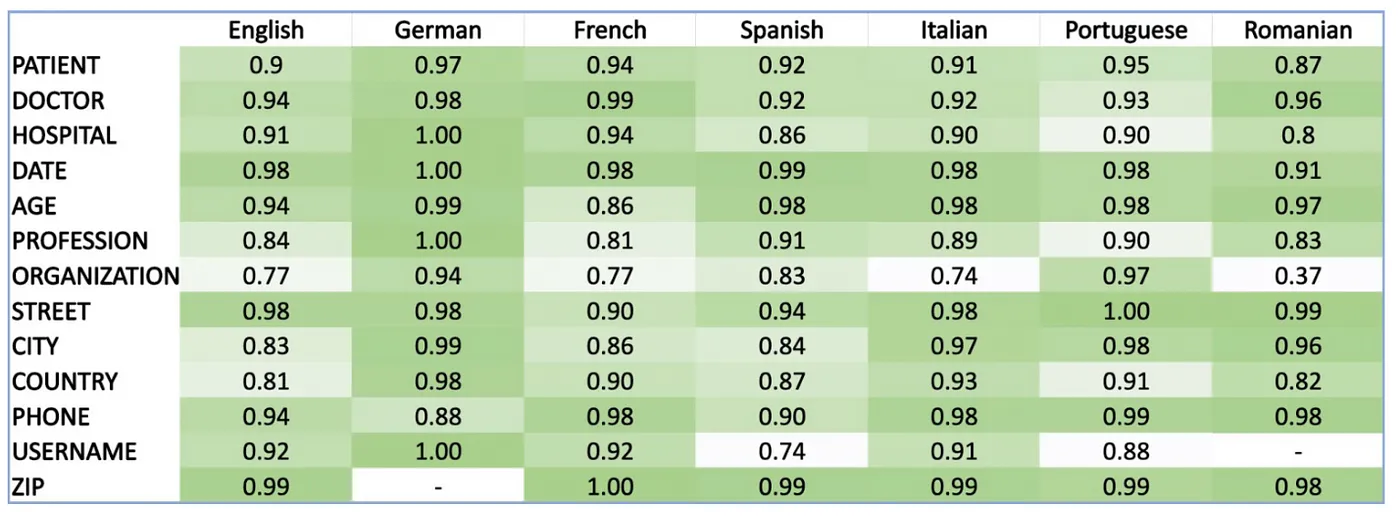
The de-identification module Spark NLP for Healthcare supports 8 languages out of the box with SOTA accuracies across more than 10 PHI entities
Comparison with ChatGPT
Study Design
- Data Selection:We selected 25 clinical discharge notes from the 2014 i2b2 Deid Challenge dataset and annotated further to reduce label errors.
- Scope: The following entities are considered during this comparison: ‘ID’, ‘DATE’, ‘AGE’, ‘PHONE’, ‘PERSON’, ‘LOCATION’, ‘ORGANIZATION’.
- Model Preparation:To prevent data leakage, we examined the dataset of the selected NER models from Spark NLP for Healthcare, retraining some models from scratch and uploading them to ModelHub for evaluation purposes.
- Prediction Collection:We ran the prompts per entity in a few shot settings, and collected the predictions from ChatGPT, which were then compared with the ground truth annotations.
- Performance Comparison: We obtained predictions from the corresponding NER models and Contextual Parsers and compared those with the ground truth annotations as well (even a single token overlapping was considered a hit).
- Reproducibility:All prompts, scripts to query ChatGPT API (ChatGPT-3.5-turbo, temperature=0.7), evaluation logic, and results are shared publicly at a Github repo. Additionally, a detailed colab notebook to run de-Identification modules step by step can be found at the Spark NLP Workshop repo.
Results
- Out of 562 sensitive entities, ChatGPTfailed to identify 227, resulting in an accuracy of approximately 60%. Among its findings, 20% were partially matched (with at least one token overlapping with the ground truth), while 41% were fully matched.
- We then processed the same documents using Spark NLP for Healthcare’s one-liner Deidentification pipeline. Out of the 562 sensitive entities, it failed to find 41, achieving an accuracy of around 93%. Of these findings, 14% were partially matched(with at least one token overlapping with the ground truth), and 79% were fully matched.
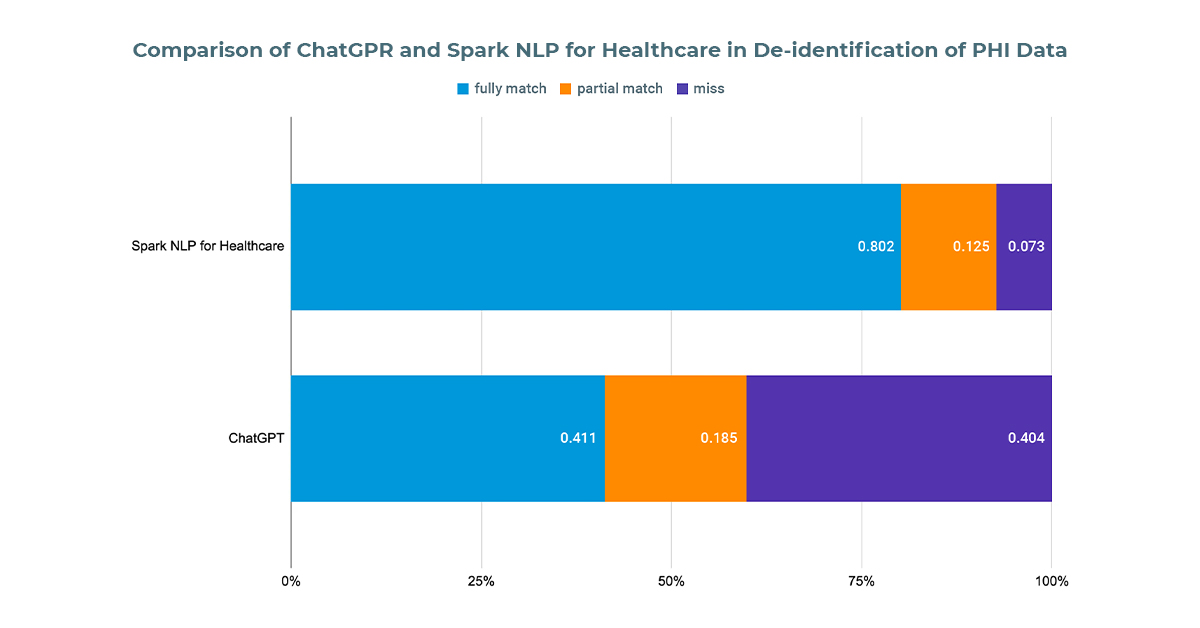
The performance of ChatGPT and Spark NLP for Healthcare in de-identifying PHI data within clinical discharge notes. The comparison includes the total number of entities, accuracy rates, and the percentage of partially and fully matched entities for both tools. Spark NLP for Healthcare demonstrates superior performance with a 93% accuracy rate compared to ChatGPT’s 60% accuracy
Conclusion
Based on the comparison, it is evident that Spark NLP for Healthcare outperforms ChatGPT in the de-identification of PHI data, with a substantially higher accuracy rate of 93% compared to ChatGPT’s 60%. Moreover, Spark NLP solutions for Healthcare demonstrates a greater rate of fully matched entities. Additionally, Spark NLP for Healthcare demonstrates a greater rate of fully matched entities.
In conclusion, although LLMs like ChatGPT offer simplicity for certain tasks, dedicated de-identification tools like Spark NLP for Healthcare provide a more reliable and accurate solution for the de-identification of sensitive data (PHI) in healthcare applications. Healthcare professionals, researchers, and organizations should consider using dedicated tools like Spark NLP for Healthcare to ensure compliance with privacy regulations and maintain the highest levels of data security and integrity.