Electronic Health Records (EHRs) hold immense potential for improving oncology care. They contain detailed histories, diagnostic findings, treatment plans, and physician notes, all of which are essential for delivering personalized cancer care. Yet much of this data remains underutilized due to its unstructured format and inconsistencies in documentation.
Natural language processing (NLP) and large language models (LLMs) offer a way forward, enabling oncology teams to extract, structure, and analyze data at scale. This blog explores how AI is transforming oncology through better EHR utilization and the tangible benefits it offers clinicians, researchers, and administrators.
The Hidden Burden: EHR Documentation Errors in Oncology
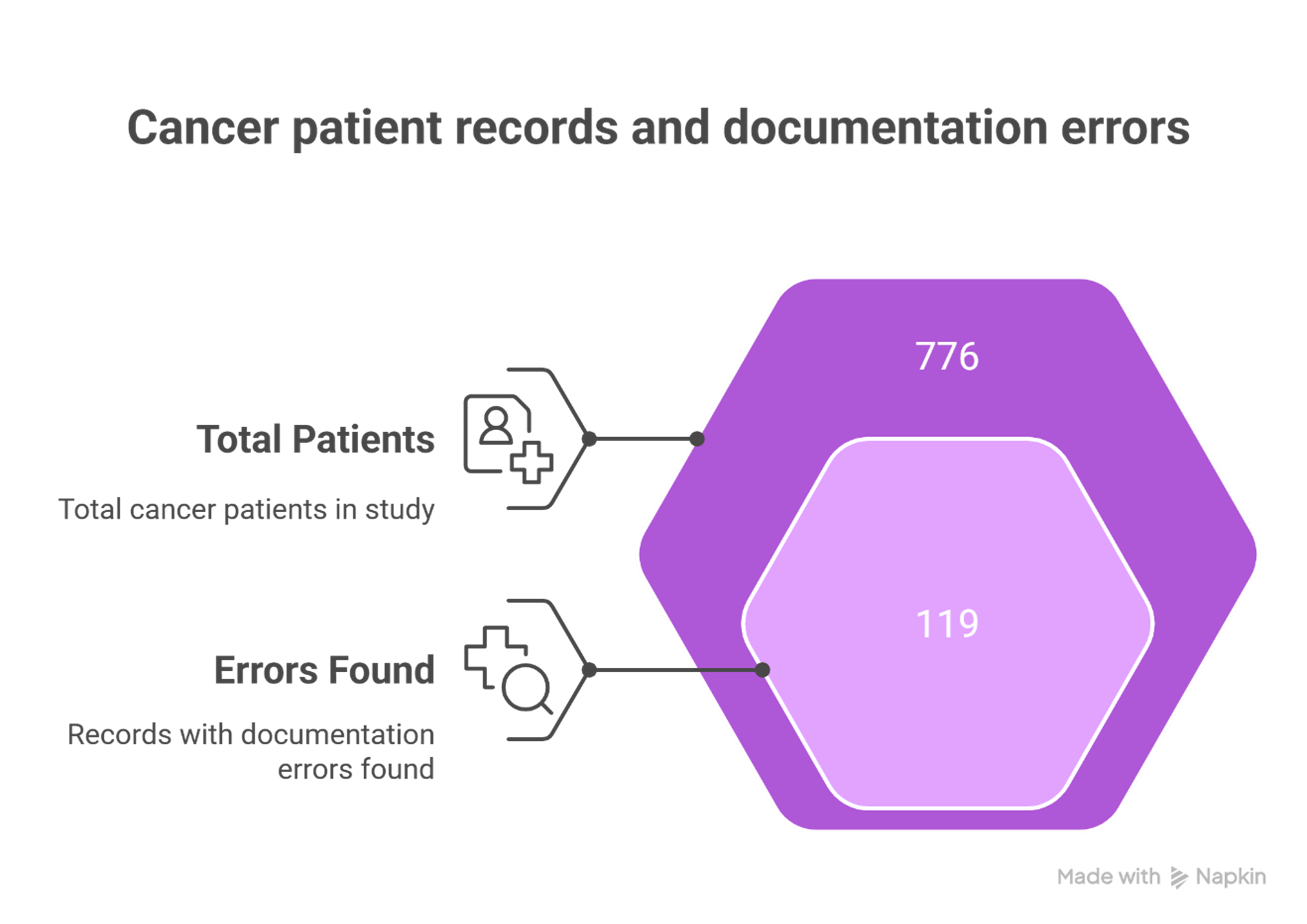
Despite best intentions, EHRs often contain errors that affect downstream decision-making. A recent real-world study examining 776 cancer patients identified a documentation error rate of 15.3%1. These mistakes can delay treatment, skew clinical research, and hinder quality reporting.
Healthcare NLP from John Snow Labs helps identify and correct inconsistencies by comparing notes across sources and surfacing anomalies. LLMs extend this by understanding medical context, flagging unlikely statements, and even suggesting plausible corrections.
Scaling Manual Abstraction: From Dozens to Thousands of Notes
Manual data abstraction is a major bottleneck in clinical trials and outcomes research. For instance, a study of 622 lung cancer patients involved reviewing over 55,000 clinical notes to extract RECIST measurements2. Scaling such efforts without automation is infeasible.
With tools like the Generative AI Lab, domain-specific LLMs dramatically accelerate this process. These models can identify key variables (e.g., tumor progression, drug regimens) and output structured data, enabling faster cohort identification and deeper insight generation.
Cancer-Specific NLP Models: From General AI to Precision Oncology
Generic LLMs often lack the granularity needed for oncology. John Snow Labs’ Healthcare NLP includes cancer-specific models trained on diverse oncology datasets, optimized for extracting tumor staging, treatment regimens, and progression metrics. These models are rigorously benchmarked to meet clinical precision needs, delivering reliable outputs that accelerate oncology workflows.
This shift from general-purpose NLP to precision-tuned models reflects a broader trend: clinical teams want models that speak the language of oncology. John Snow Labs offers Healthcare NLP pipelines fine-tuned for multiple cancer types, enhancing model performance and clinical trust.
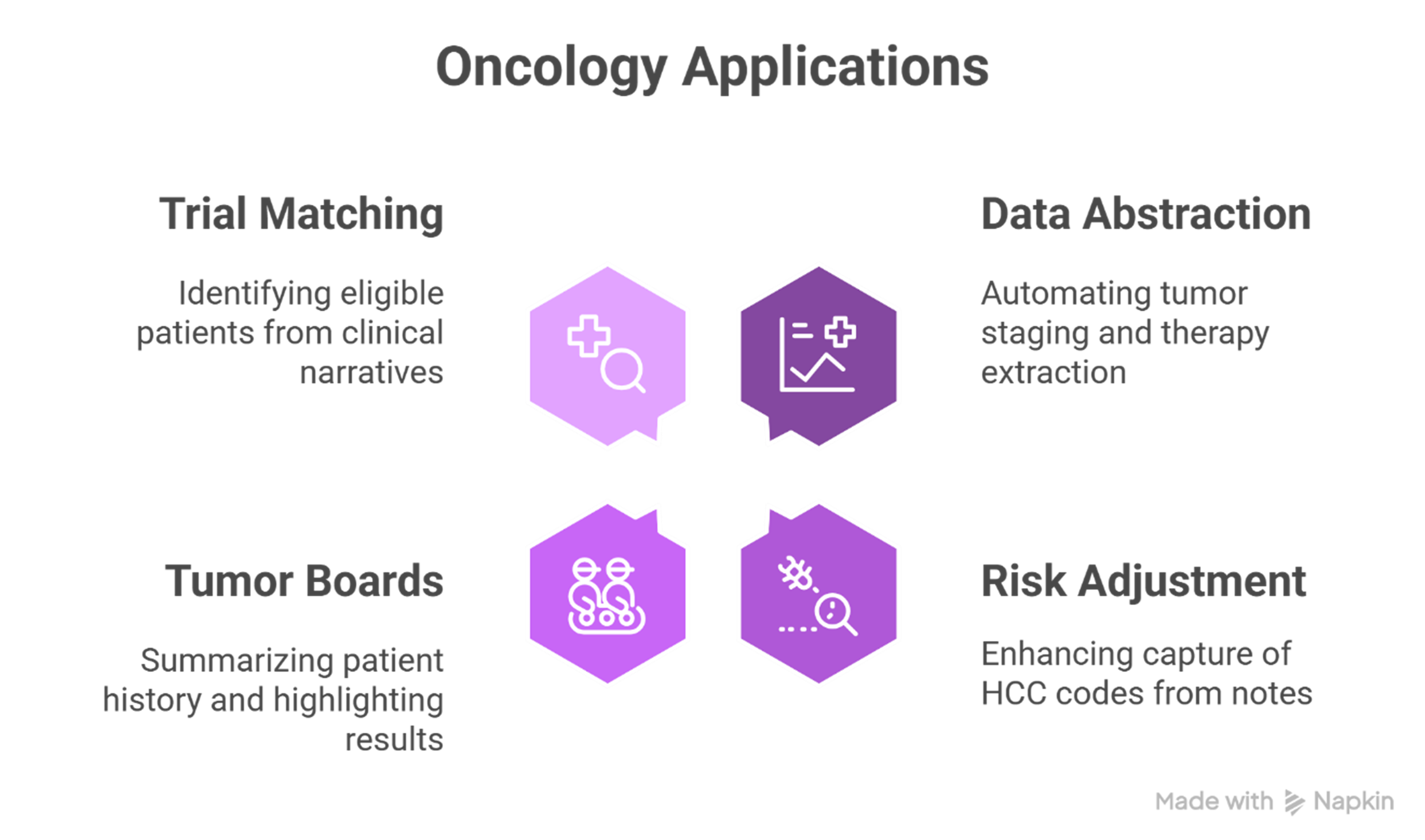
Real-World Applications: From Risk Adjustment to Tumor Board Support
NLP and LLMs from John Snow Labs are already driving impact across oncology workflows:
- Clinical Data Abstraction: Automating tumor staging and therapy extraction from radiology and pathology reports.
- Risk Adjustment: Enhancing capture of HCC codes from oncology notes to improve Medicare Advantage accuracy.
- Tumor Boards: Summarizing patient history and highlighting recent imaging or lab results.
- Trial Matching: Identifying eligible patients from unstructured clinical narratives.
These capabilities reduce administrative burden and make actionable insights available in real time.
Looking Ahead: Trust, Transparency, and Tailoring Models
As adoption grows, oncology stakeholders must ensure that NLP and LLM models remain trustworthy and transparent. This involves:
- Using explainable AI techniques to clarify model outputs.
- Continuously retraining on validated oncology datasets.
- Prioritizing compliance and auditability for clinical deployment with tools like Generative AI Lab.
The future of oncology insights lies not in replacing clinicians but in augmenting their work. By unlocking structured value from unstructured EHR data, Healthcare NLP and LLMs empower more informed decisions, timely interventions, and better patient outcomes.
To learn more about Healthcare NLP and its applications in oncology, explore our latest tools and case studies.
References
[1] H. Khela, J. Khalil, N. Daxon, Z. Neilson, T. Shahrokhi, P. Chung, and P. Wong, “Real world challenges in maintaining data integrity in electronic health records in a cancer program,” Journal of Biomedical Informatics, vol. 151, p. 104498, 2024. [Online]. Available: https://doi.org/10.1016/j.jbi.2024.104498 [2] Y. Li, Y.-H. Luo, J. A. Wampfler, S. M. Rubinstein, F. Tiryaki, K. A. V., J. L. Warner, H. Xu, and P. Yang, “Efficient and accurate extracting of unstructured EHRs on cancer therapy responses for the development of RECIST natural language processing tools: Part I, the corpus,” JCO Clinical Cancer Informatics, vol. no. 4, pp. 383–391, 2020. DOI: 10.1200/CCI.19.00147. [Online]. Available: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7265793/