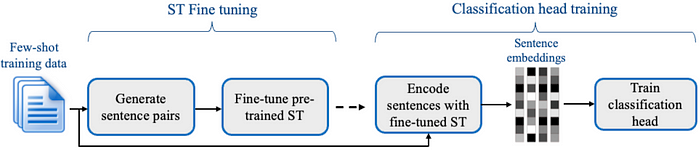
Assertion status detection is critical in clinical NLP but often overlooked, leading to underperformance in commercial solutions like AWS Medical Comprehend, Azure AI Text Analytics, and GPT-4o. We developed advanced assertion detection models, including fine-tuned LLMs, transformers, few-shot classifiers, deep learning (DL) and rule -based approaches. Our fine-tuned LLM achieves 0.962 accuracy, outperforming GPT-4o (0.901) and commercial APIs, with notable improvements in Present (+4.2%), Absent (+8.4%), and Hypothetical (+23.4%) assertions. Our DL models also excel in Conditional (+5.3%) and Associated with Someone Else (+10.1%) categories. The few-shot classifier (0.929 accuracy) provides a lightweight alternative for resource-limited environments. Integrated with Spark NLP, our models offer scalable, transparent, and domain-adapted solutions that surpass black-box commercial APIs in medical NLP tasks.
Introduction
The adoption of Electronic Health Records (EHRs) has revolutionized healthcare, creating vast repositories of patient data. To extract actionable insights from these records, Natural Language Processing (NLP) plays a crucial role, particularly in assertion detection, which determines whether a medical concept is present, absent, possible, hypothetical, conditional, or associated with someone else.
Early assertion detection relied on rule-based methods like NegEx and ConText, which achieved high precision but struggled with recall. Deep learning and transformer-based models have since emerged, offering improved contextual understanding but facing challenges like class imbalance and data requirements.
In this blog post, we introduce a comprehensive assertion detection framework integrated into Spark NLP, leveraging LLMs, deep learning, transformers, few-shot learning, and rule-based methods. Our approach surpasses traditional negation detection by addressing the full spectrum of assertion types, ensuring scalable, accurate, and efficient clinical NLP applications.
Example Pipeline
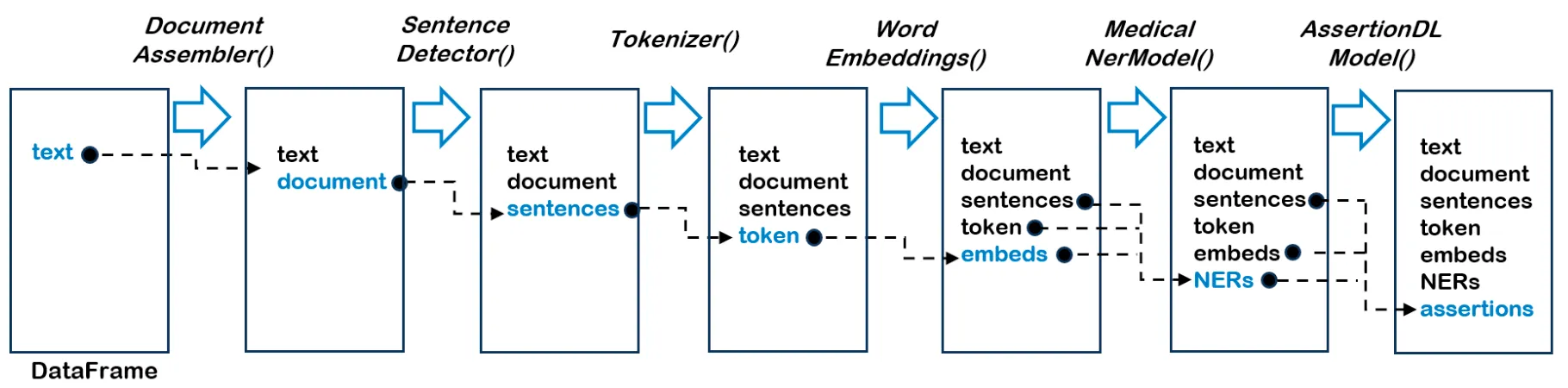
The flow diagram of a Spark NLP pipeline. When we fit() on the pipeline with a Spark data frame, its text column is fed into the DocumentAssembler() transformer and a new column document is created as an initial entry point to Spark NLP for any Spark data frame. Then, its document column is fed into the SentenceDetector(), Tokenizer() and WordEmbeddings(). Now data is ready to be fed into NER models and then to the assertion model
The flow diagram of a Spark NLP pipeline. When we fit() on the pipeline with a Spark data frame, its text column is fed into the DocumentAssembler() transformer and a new column document is created as an initial entry point to Spark NLP for any Spark data frame. Then, its document column is fed into the SentenceDetector(), Tokenizer() and WordEmbeddings(). Now data is ready to be fed into NER models and then to the assertion model.
Dataset and Label Description
Our benchmarking was conducted on the official 2010 i2b2 dataset (test split), a standard resource for assessing assertion detection in clinical NLP. The dataset includes six assertion categories: Absent, Associated with Someone Else, Conditional, Hypothetical, Possible, and Present.
To enhance fine-tuning efficiency, our fine-tuned LLM excludes the Conditional label, as its overlap with Hypothetical could introduce ambiguity. However, other models, including LLMs, retain all six categories to ensure a comprehensive performance evaluation across all assertion types.
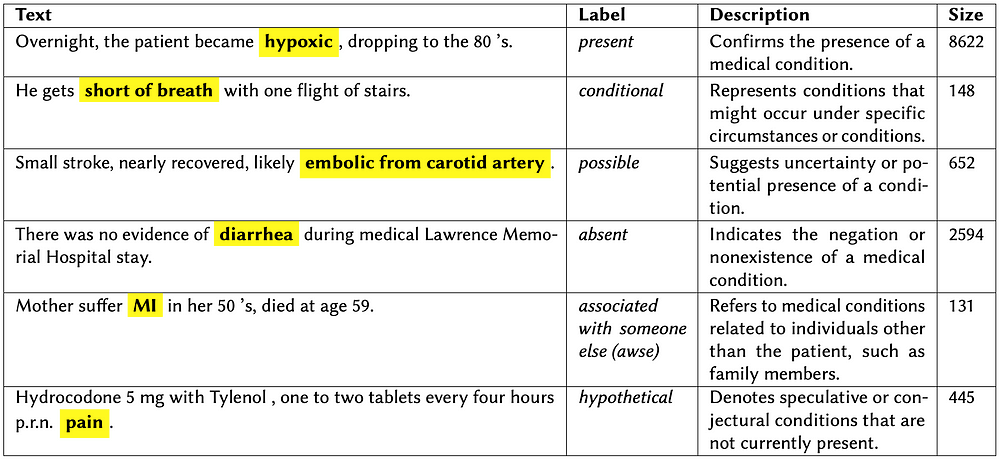
Examples from the i2b2 dataset illustrating different assertion classes with their label distribution.
Assertion Detection with LLMs
Traditional assertion detection methods struggle with rare assertion types and require manual rule creation. To enhance accuracy, we fine-tuned LLama-3.1–8B on the i2b2 dataset using LoRA fine-tuning. This approach reduces memory overhead while preserving pre-trained knowledge. Key optimization include:
- Using a context windowing strategy (two sentences before and after the target text).
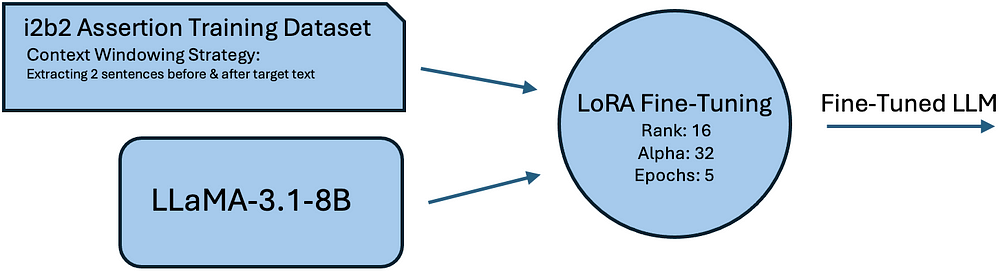
The illustration shows the fine-tuning process of LLaMA-3.1–8B for assertion detection, from i2b2 dataset preprocessing to LoRA optimization.

Example of Fine-tuned LLM prompt for detecting assertion status in medical records
Assertion Detection via DL Model
We developed AssertionDL, a Bi-LSTM-based model that processes medical entities alongside their surrounding context. By analyzing i2b2 data, we optimized a context window of 9 tokens left, 15 right, balancing performance and efficiency. Integrated into Spark NLP, this model provides scalable and accurate assertion detection.
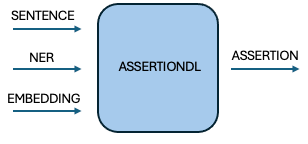
An illustration depicting the inputs and outputs of AssertionDL in a pipeline.
Assertion Detection via BERT for Sequence Classification (BFSC)
Rather than generating new text, BFSC treats assertion detection as a classification task. We fine-tuned BioBERT on medical text, ensuring better focus on target entities. Experimenting with different context sizes showed minimal accuracy gains while increasing processing time, making a standardized context size the best approach.
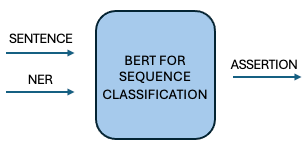
An illustration depicting the inputs and outputs of BertForSequenceClassification in a pipeline
Few-Shot Assertion Detection via Transformers
Our FewShotAssertion model, based on SetFit, enables high accuracy with minimal labeled data. Using sentence-transformer embeddings and contrastive learning, it efficiently aligns assertion categories, making it ideal for resource-limited scenarios.
SetFit model training architecture. Image from the open-source Arxiv paper: https://arxiv.org/abs/2209.11055
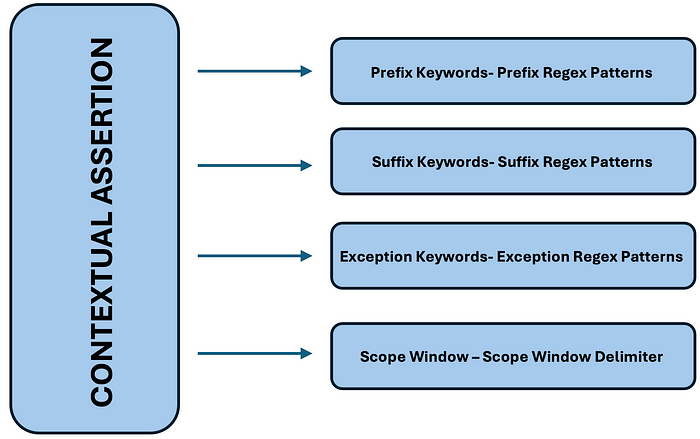
Rule-Based Assertion Detection with Contextual Awareness
Our ContextualAssertion model expands rule-based detection with customizable keyword sets, regex patterns, and scope windows. Unlike fixed-pattern models, it provides greater flexibility and adaptability for complex medical texts.
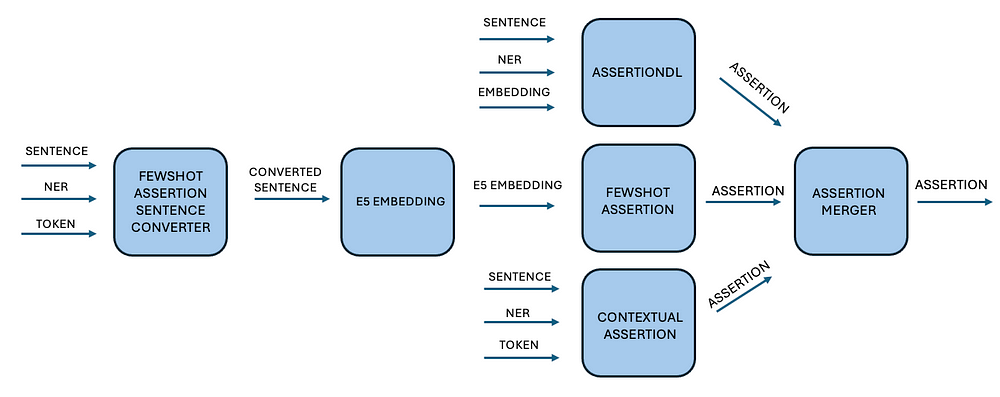
Integrating Assertion Detection into Healthcare NLP
To enable end-to-end assertion detection, we developed a pipeline that:
- Extracts named entities using NER models.
- Uses a stacking approach (DL, few-shot, and rule-based models) to improve accuracy. (Stacking approach is an ensemble learning method that combines multiple models to improve overall accuracy.)
- Applies a majority voting mechanism to resolve prediction conflicts.
This robust pipeline, integrated within Spark NLP, delivers scalable and high-performing assertion detection for clinical NLP applications.
Benchmarking Assertion Detection Models
To evaluate the performance of assertion classification, we benchmarked our models — AssertionDL, FewShotAssertion, ContextualAssertion, and BFSC — against industry-standard solutions such as NegEx, AWS Comprehend Medical, Azure AI Text Analytics, and GPT-4o.
NegEx, a rule-based system, detects negation in clinical text by identifying predefined linguistic patterns. While effective for simple negation, it lacks flexibility for more nuanced assertion types.
GPT-4o was tested on the i2b2 dataset with a carefully designed prompt to classify assertion statuses. Due to data-sharing restrictions, we obfuscated the dataset using John Snow Labs’ Healthcare NLP tools before running evaluations.

Example of GPT-4o prompt for detecting assertion status in medical records
AWS Comprehend Medical and Azure AI Text Analytics extract medical entities and assign assertion labels (e.g., present, absent, hypothetical). Since these services only classify entities they extract, we evaluated them on partially or fully overlapping entities with the i2b2 dataset. To ensure consistency, assertion labels were mapped to i2b2 equivalents, and matches were categorized as Full Match, Partial Match, or No Match.
Our evaluation highlights the limitations of rule-based and black-box commercial solutions while demonstrating the advantages of domain-adapted, transparent, and customizable assertion detection models for clinical NLP.
Performance and Practicality of Assertion Detection Models
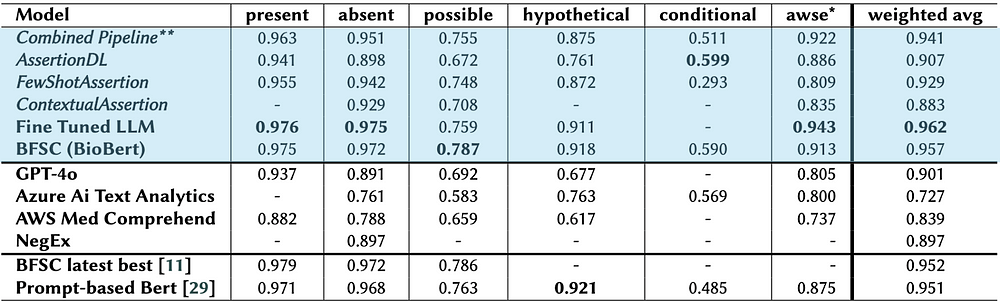
Comparison of assertion models across various categories. Best performing model for each category is represented with bold characters. The models in the first section of this table are developed by JSL. In LLM and GPT-4o experiments, hypothetical and conditional labels are merged/treated as a single label.
Our fine-tuned LLM (LLaMA 3.1–8B + LoRA on i2b2) achieves top-tier performance across most assertion categories, surpassing GPT-4o, Azure AI, and AWS Comprehend, with minor gaps in possible and hypothetical labels. This underscores the power of domain-adapted models with well-crafted prompts.
Our hybrid pipeline, integrating rule-based and machine learning models, performs on par with the LLM while outclassing commercial solutions, particularly due to customization and fine-tuning options.
For flexibility, individual components offer strong results:
- AssertionDL excels in conditional and associated with someone else labels, outperforming GPT-4o.
- FewShotAssertion is ideal for resource-constrained settings, contributing to absent and hypothetical categories.
- BFSC (BioBERT) benefits from domain adaptation, rivaling AssertionDL in performance.
Despite its accuracy edge, LLM-based detection is 100× slower and thousands of times more expensive than our deep-learning models, making our lightweight, domain-adapted solutions the most scalable and cost-effective option for clinical NLP applications.
Speed Performance Comparison:
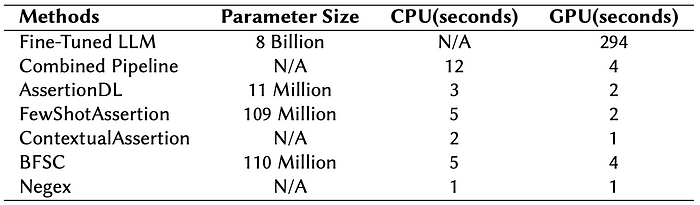
Mean latency per 100 rows, measured in seconds for various assertion methods. Experiments were run on Google Colab servers, with CPU tasks performed on a CPU instance (8vCPU @ 2.2 GHz, 50.99 GB RAM) and GPU tasks executed on an NVIDIA A100 GPU (40 GB HBM2).
Label Correspondences:

AWS Comprehend Medical model does not represent conditional labels. Azure AI Text Analytics for Health has 3 distinct possible labels — positive, negative and neutral, while present is not represented.
Pre-trained Assertion Models in Healthcare NLP:
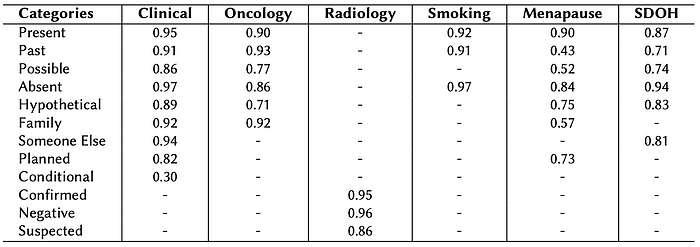
F1 Score Comparison of pre-trained models across various assertion categories. Each row corresponds to a specific pre-trained model — such as Clinical, Oncology, Radiology, Smoking, Menopause, and Social Determinants of Health (SDOH) — highlighting their strengths in different assertion labels. Overall, this comparison serves as a valuable resource for practitioners and researchers seeking to select the most appropriate model for their specific application in medical text analysis.
Conclusion: Scalable & Accurate Assertion Detection
We evaluated JSL’s assertion detection models, from lightweight DL models to fine-tuned LLMs. Our fine-tuned LLM achieved 0.962 accuracy, outperforming GPT-4o (0.901) and commercial APIs, especially in Present, Absent, and Hypothetical assertions. However, it is 100× slower and thousands of times more expensive, making it impractical for scalable clinical NLP.
Our DL-based models (AssertionDL, FewShotAssertion, BFSC) provide high accuracy with superior efficiency, excelling in key assertion categories. The Combined Pipeline outperforms all commercial solutions, offering a balance of accuracy, speed, and cost-effectiveness.
Integrated into Spark NLP, these pre-trained assertion models deliver production-ready, scalable alternatives to black-box solutions like GPT-4o, Azure AI, and AWS Comprehend, ensuring accurate medical insights in real-world applications.
Notebook below includes benchmark results obtained using the official i2b2 dataset. Due to privacy restrictions, we are unable to share the dataset itself — however, you can still review the performance metrics derived from it.
Additionally, the notebook showcases example assertion outputs from the Combined Assertion Pipeline on a sample clinical text.
Check out the Combined Assertion Pipeline here
How to Get Access & Stay Updated
John Snow Labs models are licensed, so if you want to use these models, you can watch “Get a Free License For John Snow Labs NLP Libraries” video and request one from the John Snow Labs web page.
You can follow us on medium and Linkedin to get further updates or join slack support channel to get instant technical support from the developers of John Snow Labs. If you want to learn more about the library and start coding right away, please check our certification training notebooks.