Integration and interoperability are becoming very common terms for anyone working in the IT healthcare field. You can find them among the requirements of any job or in any health informatics academic curricula or online courses.
Integration and interoperability are so important to the extent that made many organizations and scientists exerts years of effort to set definitions and standards for it. Their main target was to make all vendors a standardized communication format.
But, why are they so important and is there a difference between integration and interoperability?
Although people use the 2 words interchangeably, there is a great difference between them.
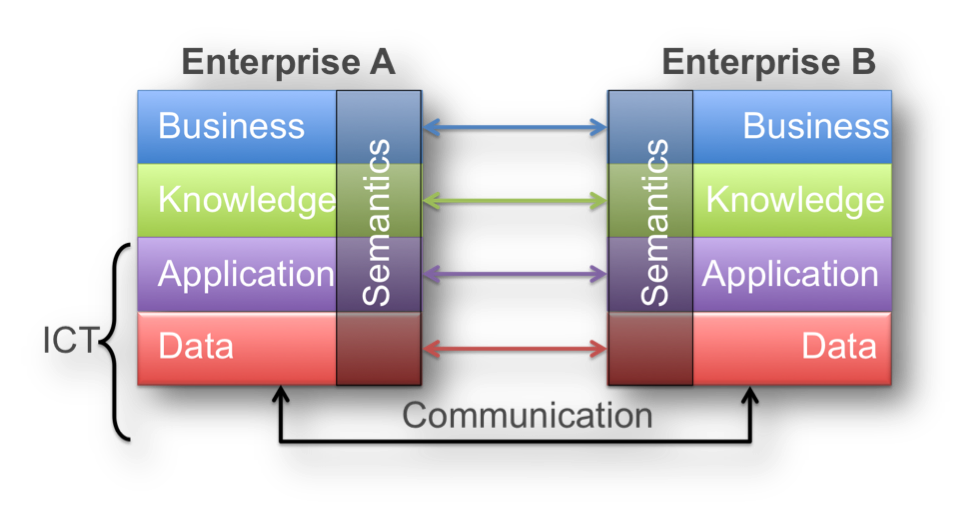
Integration is a process where a hardware device and a software application can send and receive data efficiently (like an ECG device and an EMR system for example).
Interoperability, on the other hand, is concerned with connecting patients, medical devices with different management systems or software applications, to produce a new context where clinical and administrative insights can be obtained as a result of combining different data from different resources.
The importance of integration and interoperability can be felt more in the healthcare sector due to the fragmented nature of the services offered.
In any healthcare facility, different services are offered through different departments (admission, diagnosis, laboratory and investigation work, radiology, surgeries, pharmacies, billings, … etc).
All the healthcare workers in the same facility must have instant and secure access to all (or most) of these data at the same time. In addition, any updates done by any service provider must lead to a synchronized update in the patient file or the database, which can be seen at the same time by any other service provider or decision maker.
Another scenario may include an admitted patient in an intensive care department lying on a bed that has more than tend devices attached. The clinician may need to analyze combined data from these devices to take the final decision on a specific treatment plan.
The devices must have a unified communication standard both on the semantic and the technical level.
These complex needs and entangled organizational structure need a unique governance model, where a fully integrated and interoperable system is available for decisions makers.
Obtaining such hermetic interoperability should face different challenges. According to the nature of these challenges, we can divide interoperability into 3 levels:
- Foundational Interoperability: to what extent the system can send/receive messages successfully.
- Structural Interoperability: to what extent the system can recognize the data fields (with no reference to contents).
- Semantic Interoperability: to what extent the system can understand the data fields and their contents
The start was with The Certification Commission for Health Information Technology (CCHIT) who set a definition to the most important specifications for EMRs with respect to three factors (interoperability, security, and functionality).
Another definition was developed by the Healthcare Information and Management Systems Society (HIMSS). HIMSS defined interoperability as “In healthcare, interoperability is the ability of different information technology systems and software applications to communicate, exchange data, and use the information that has been exchanged.”
The Office of the National Coordinator for Health Information Exchange (ONC) developed 16 elements which represent the “Common Meaningful Use Data Set”. This is simply a standard set of data. Moreover, (ONC) developed its own framework which is composed of four components (vocabulary/code sets, content structure, transport, and services).
More and more scientists and organizations realized the importance of developing standardized semantics according to the nature of their field or research project.
The American College of Radiologists (ACR) and the national electrical manufacturer association developed DICOM standards to ensure proper handling of imaging data.
A group of pathologists developed the Systematized Nomenclature of Pathology (SNOP) which was then merged with the Read Clinical Term. The final standard was known as Systematized Nomenclature of Medicine – Clinical Terms (SNOMED-CT).
The FDA, the Veterans Administration (VA), the U.S. National Library of Medicine (NLM), and the pharmacy knowledge base vendors developed the RxNorm (a part of the Unified Medical Language System (UMLS)). RxNorm is specific to medication and medical allergies. It maps NDC codes with the VA’s National Drug File (VANDF) and other drug terminologies.
Clement J. McDonald and Stanley M. Huff. LOINC developed the Logical Observation Identifiers Names and Codes (LOINC) which is concerned with names and codes for laboratory tests, radiology exams, instruments used and observations.
The World Health Organization (WHO) developed the different revisions for the International Classifications of Diseases, which is used to report different patient diagnoses.
Semantic interoperability
There are other causes which might enforce the use of semantic interoperability. The idea of using data from discrete EMR systems or research network to recruit patients who are compliant with the pre-defined inclusion criteria enforces the need for semantic interoperability. The evolution of Clinical Trial Recruitment Support Systems (CTRSSs) reduced the costs of the clinical trials to a great extent because most of the costs of clinical trial were consumed in the recruitment process. The success of CTRSSs depends mainly on proper semantic interoperability.
For example, MI could be interpreted as a “Myocardial Infarction” in a system used by a research institute while being interpreted as “Mitral Insufficiency” in another.
Clinical decisions also need fully interoperable data, as data may come from different servers or systems. A physician may need to revise a radiology report (from the RIS) and have a look on the Complete Blood Count (CBC) (from the LIMS) before deciding the final treatment plan suitable for a specific case.
Researchers, clinicians, software houses dealing with medical applications may be in continuous need for different medical terminology datasets. Such datasets are not abundant on the internet and if they are found, most of the time they are not accurate.
John Snow Labs catalogs have more than 1775 normalized datasets, most of them are freshly curated and machine and manually validated. In this catalog, you can find curated datasets for SNOMED Clinical Terminology, RxNorm and many other curated datasets for standardized medical terminologies.
Using such curated datasets may save up to 80 % of the researcher time. It is reported now that the cleaning and preparing data process may consume up to 80% of the data scientist time.
The availability of a curated and well-revised interoperable semantics and terminologies will save time in another way as the researcher will not waste time thinking which term to use or which is better.
Usage of such standardized medical terminologies is not only restricted to research. It can also be used in governmental reporting (reporting morbidity and mortality) or in monitoring systems (for reporting adverse reactions).
Ensuring hermetic data integration and interoperability is crucial for maximizing the value of healthcare data. By employing solutions that leverage Generative AI in Healthcare and Healthcare Chatbot technologies, organizations can enhance data usability, improve patient outcomes, and streamline healthcare operations across various platforms.