The emergence of new artificial intelligence concepts like face recognition, supervised-learning science, and predictive analytics have imposed new challenges to medical imaging.
In addition, the emergence of new imaging techniques has yielded a need for huge storage capacities and appropriate big data analytics techniques. Histology studies can reach hundreds of megabytes while CT scans slices can exceed 2500 images per study.
To make use of the stored images, a precise and fast supervised learning algorithm is needed.
The diagnosis and prediction processes in clinical decision support applications pass through different phases. Labeling can be considered the initial phase in such systems that are based on medical imaging in addition to another data from other systems may be used (like laboratory information systems (LIMS)) to reach a diagnosis.
LABELING: A NEW OUTSOURCING/FREELANCE OPPORTUNITY
Many people wonder how can just taking hundreds or thousands of images can help in reaching a diagnosis. Of course, this is not done through traditional algorithms or through traditional programming techniques.
It was not until the emergence of artificial intelligence algorithms and the concepts of supervised learning, until the ability to reach a precise prediction or a definitive diagnosis.
Supervised learning maps an input to an output based on the given labeled training data supplied.
There are 2 types of supervised learning: Regression and Classification.
Classification identifies the category to which a new data point belongs, based on the supplied training dataset.
Regression is concerned with finding the relations between the input variable from the training dataset, in order to be able to predict the outcomes of new events.
Supervised learning algorithms are applied to previously labelled images.
One of the commonly used websites that could help in the labeling process is www.labelbox.com.
Labelbox is the best tool that can be used to annotate medical imagery within the scope of HIPAA regulations. It can help SMEs to build custom applications on top of Labelbox.
The following is an example that can help the reader to understand how the process is done:
If we are developing a system to diagnose the diseased tongue (like fissured tongue or geographic tongue), you must upload many thousands of images of healthy and diseased tongues to a tool like Labelbox and label the healthy areas and the diseased ones. These labeled images will be considered the training dataset that will be used to develop a supervised machine learning algorithm to detect diseased cases.
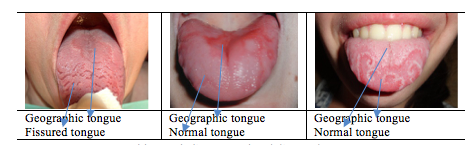
Table 1: Labeling normal and diseased tongue
Labeling is becoming a great freelance and Business Process Outsourcing (BPO) opportunities. Job seekers with good healthcare educational background can find a lot of jobs on Upwork and freelancer or different freelance websites. You can make use of your knowledge by just labeling healthy/diseased areas on the images.
We can imagine the virtue of such methodology on the detection of premalignant lesions and accordingly early detection of oral cancer (and other types of malignant tumors).
The whole process can be described as “Ontology-based diagnostic decision support”.
May be some readers can’t realize what is meant by the term “Ontology”.
ONTOLOGY
Ontology is the study of the nature of being, categories of being and their relations. It is concerned with studying what entities exist and finding the optimum to group them and find any probable relation to be able to arrange them in a hierarchical form.
2.1 Ontologies in Radiology
Different types of ontologies exist. Most of them are available here.
In 2006, the RSNA initiated a project aiming to create a radiology-specific ontology. The end-product of the project was RadLex;a set of radiology terms used in radiology reporting, decision support, data mining, and data registries.
The Radiation Oncology Ontology(ROO) main aim is to include the whole radiation oncology domain while considering the existing ontologies.
2.1.1 Ontology Components
- Concepts and Instances
May be also called elements, entities, or classes. Concepts can be defined as “unit of thoughts”.Those who are implicitly defined are called “Primitive ” while those who need relations or constraints to explicitly define them are called “defined concepts”.
Concepts are considered like classes. Hence, they can have instances.
For example, the concept Patient Name can have the instance “Michael Jordan”.
- Relations
Relations define how pairs of concepts can be related.
- Restrictions (Axioms)
Restrictions or Axioms are logical statements attached to concepts and explicitly define it.
- Inheritance
The mechanism where child concepts inherit all the properties of the parent concept.
2.1.2 Types of Ontologies
- Upper-Level Ontology
A Domain-independent ontology that represents basic or primitive concepts and relations. The main aim of it is to ensure integration and interoperability of the domain-specific ontologies.
- Reference Ontology
With the incremental increase in ontologies standards available on the web, it becomes a mandate to develop a Reference ontology that suits different knowledge domains.
For example, the Common Anatomy Reference Ontology (CARO) aims to enforce the interoperability between different available anatomy ontologies for different species.
Another example is the Foundation Model of Anatomy (FMA), an ontology of structural human anatomy.
- Application Ontology
Application ontologies are commonly used if there is a need to move from a domain to another. An application ontology should be tested against pre-defined use cases and competency scenarios. It is different than Upper-level Ontology in that it targets a specific group to perform a specific task and a specific well-defined area of knowledge.
2.1.3 Application Area of Ontologies in Radiology
- American College of Radiology Appropriateness Criteria (ACRAC)
During 1990, ACR developed criteria for the appropriate use of medical imaging. These criteria include imaging appropriateness for specific clinical problems depending on the patient medical history and condition, benefits versus exposure to harmful radiation.
The Centers for Medicare & Medicaid Services (CMS) receives the claims from different facilities and hospitals which treat Medicare patients. CMS analyze them to assess the use of medical imaging.
John Snow Labsdatabase repository can be considered a good resource where you can obtain data about the Outpatient Imaging Efficiency Core Measures by Hospitalor by state.
Both datasets include the hospital data for the Outpatient Imaging Efficiency Core Measures. These Core Measures gives information about the hospitals’ use of medical imaging tests for outpatients. Examples of medical imaging tests include CT Scans, MRIs, and mammograms.
- Best Practices Guidelines
Many trials were done to reach the ideal use of medical imaging that matches each specific case.
Different efforts were done towards achieving the optimum guidelines.A computable paper-based clinical guideline called GEODE-CM system for guidelines and data entry emerged in 1990.While GEODE-CM system focuses on guidelines and data entry, the MBTA system focuses on medical logical modules for alerts and reminders.
The EON Architecture and the PRODIGY system guideline-based decision support are among the most remarkable efforts to be mentioned here.
All these guidelines were different in format and functionality, therefore there was a great need for a common line format.
In 1998, the guideline interchange format (GLIF); a sharable computer-interpretable clinical practice guideline was developed.
Former functionalities from previous systems were involved in addition to 3 abstraction levels:
-
- A conceptual level (human-readable) for medical terms represented as free text in flow charts;
- computable level that execute guidelines;
- and an implementation level to integrate guidelines in institutional clinical applications.
The integration between appropriate medical imaging criteria and imaging results to develop computer-interpretable clinical guidelines is possible providing the presence of interoperability between the two systems.
Hermetic interoperability needs accurate data presented in a coherent and standardized format.
The next blog coming in the next few weeks will expand the topic further to include more information for the readers interested in the multidisciplinary area that gathers medical imaging, ontology, and big data.
The integration of big data in medical imaging presents significant challenges, including data management, quality control, and the need for advanced analytical tools to derive meaningful insights. By embracing solutions such as Generative AI in Healthcare and employing a Healthcare Chatbot, healthcare organizations can address these challenges effectively, enhance diagnostic accuracy, and foster improved communication between providers and patients.






























