Spark NLP for Healthcare 2.7 is out and available now! If you’re an existing customer, expect an email with upgrade instructions. If you’re not, start your free trial here. The biggest feature in this release is Text to SQL – which is big enough to deserve its own post. Here are the other major additions in this release:
More Accurate Entity Resolution
In addition to ChunkEntityResolver, you now also have at your fingertips a suite of BioBert-based entity resolvers using the brand new SentenceEntityResolver annotator. The new annotator is fully trainable and comes with several pretrained entity resolvers for these medical terminologies:
- CPT: biobertresolve_cpt
- ICDO: biobertresolve_icdo
- ICD10CM: biobertresolve_icd10cm
- ICD10PCS: biobertresolve_icd10pcs
- LOINC: biobertresolve_loinc
- SNOMED_CT (findings): biobertresolve_snomed_findings
- SNOMED_INT (clinical_findings): biobertresolve_snomed_findings_int
- RXNORM (branded and clinical drugs): biobertresolve_rxnorm_bdcd
Example:
text = 'He has a starvation ketosis and type two diabetes mellitus'
df = get_snomed_codes (light_pipeline_snomed_int, 'snomed_code', text)
Output:

Check out the Colab notebook to see more examples and run on your data.
You can also train your own entity resolver using any medical terminology – like MedDRA or your organization’s custom ontologies. Check out this notebook to learn more about training from scratch.
More Accurate Clinical Named Entity Recognizers
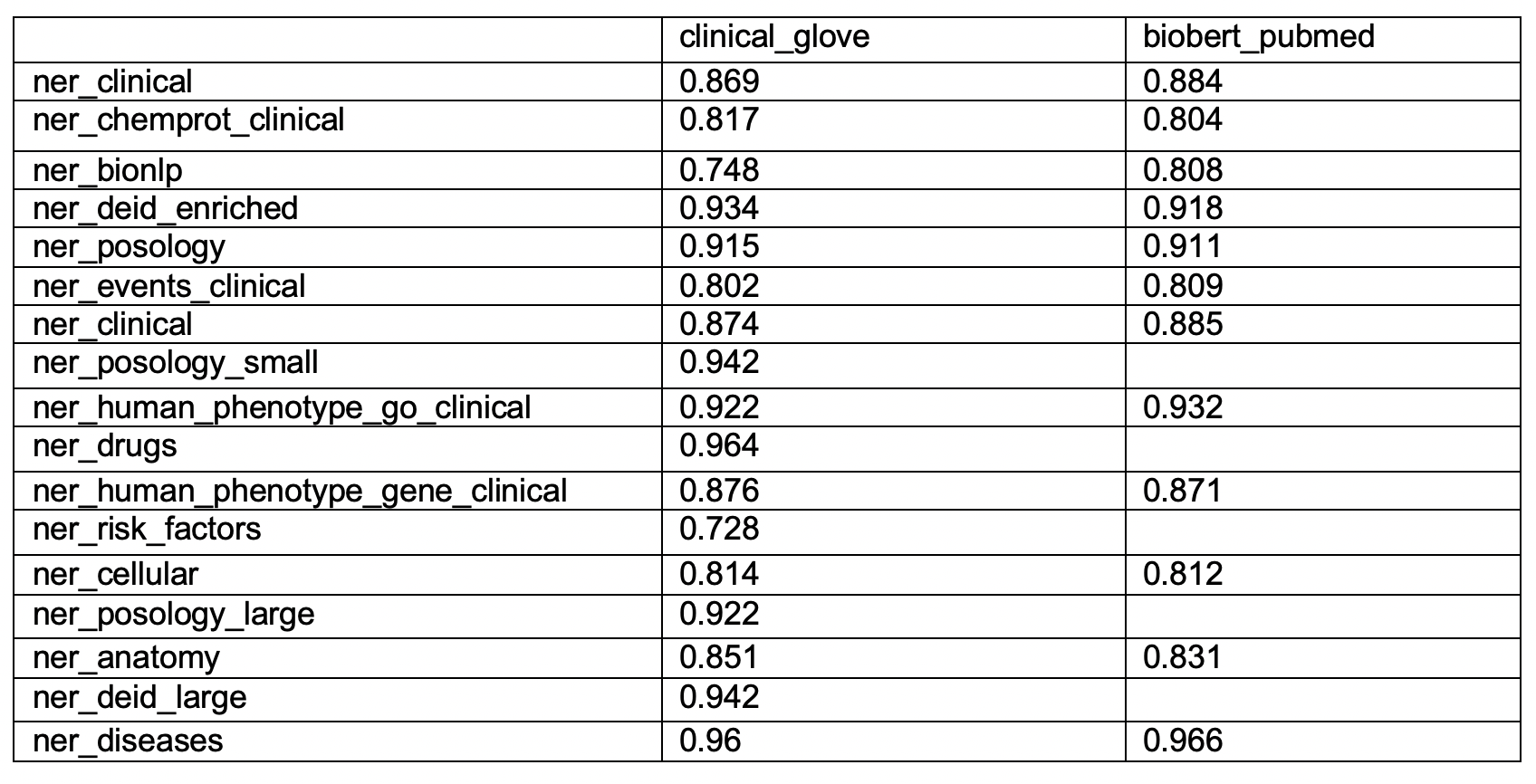
We have retrained, fine-tuned, and released new BioBERT-based versions of these 13 clinical named entity recognition models:
- ner_clinical_biobert
- ner_anatomy_biobert
- ner_bionlp_biobert
- ner_cellular_biobert
- ner_deid_biobert
- ner_diseases_biobert
- ner_events_biobert
- ner_jsl_biobert
- ner_chemprot_biobert
- ner_human_phenotype_gene_biobert
- ner_human_phenotype_go_biobert
- ner_posology_biobert
- ner_risk_factors_biobert
Metrics (micro averages excluding O’s):
New PICO Classifier for Evidence-Based Medicine
Successful evidence-based medicine (EBM) applications rely on answering clinical questions by analyzing large medical literature databases. In order to formulate a well-defined, focused clinical question, a framework called PICO is widely used, which identifies the sentences in a given medical text that belong to these four components:
Participants/Problem (P) (e.g., diabetic patients)
Intervention (I) (e.g., insulin)
Comparison (C) (e.g., placebo)
Outcome (O) (e.g., blood glucose levels).
Spark NLP now has a pretrained PICO Classifier that is trained with BioBERT embedding.
Example:
text = “There appears to be no difference in smoking cessation effectiveness between 1mg and 0.5mg varenicline.” pico_lp_pipeline.annotate(text)['class'][0] >> “CONCLUSIONS”
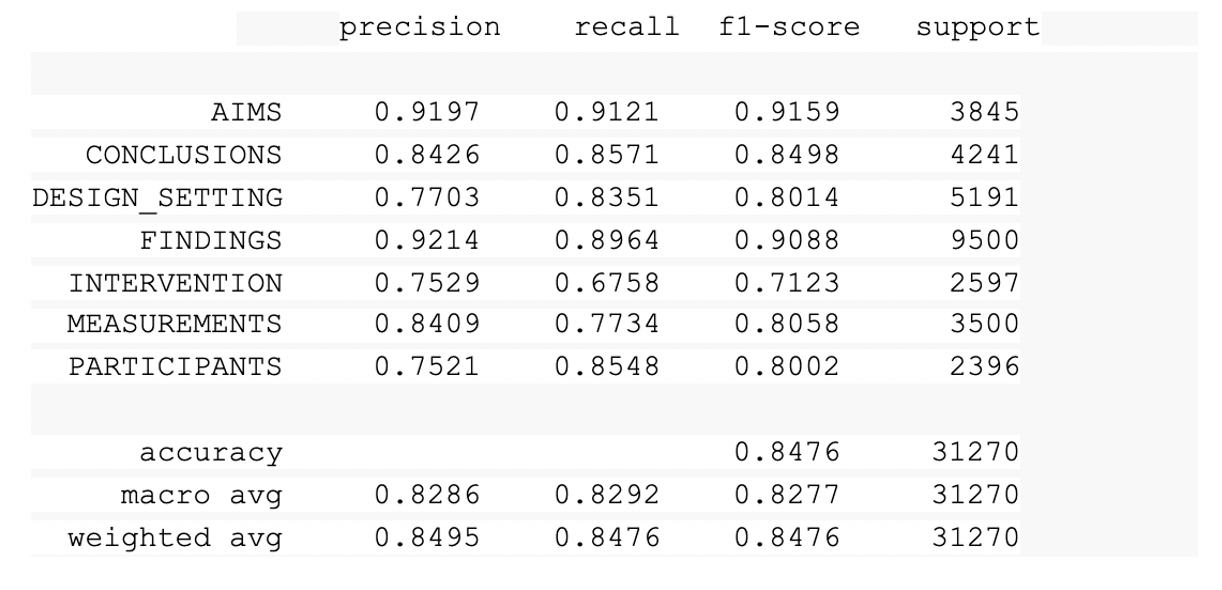
Metrics:
New Biomedical Named Entity Recognizers
We have released three new NLP biomedical models for NER, trained with clinical embeddings. All of these are single entity models:
- ner_bacterial_species – comprising of Linneaus and Species800 datasets)
- ner_chemicals – general purpose and biochemicals, comprising of BC4Chem and BN5CDR-Chem)
- ner_diseases_large – comprising of ner_disease, NCBI_Disease, and BN5CDR-Disease
New Clinical & Traffic Accident NER Models in German
Two new NER models for the German language have been trained, optimized, and released:
- ner_healthcare_slim
('TIME_INFORMATION',
'MEDICAL_CONDITION',
'BODY_PART',
'TREATMENT',
'PERSON',
'BODY_PART')
Check out this Colab notebook to test these with other German clinical NLP models (clinical NER and ICD10GM).

- ner_traffic
Extract entities related to traffic accidents like date, trigger, location, and more.

Check out this Colab notebook to test further.
Support for Overlapping Entities
In order to use multiple NER models in the same pipeline, Spark NLP includes the ChunkMerge Annotator which is used to return entities from each NER model by overlapping. Now, this annotator has a new parameter to avoid merging overlapping entities (setMergeOverlapping) to return all the entities regardless of character indices.
This is useful to analyze what every NER module returns on the same text – when you are using multiple NER models in the same NLP pipeline or comparing the output of competing NER models.
The online documentation has been updated with this new release and the Colab notebooks are current too. Give it a go – and let us know what you think and what we should build next.
As Generative AI in Healthcare continues to evolve, innovative applications such as the Healthcare Chatbot are revolutionizing patient interactions, providing real-time support, and assisting healthcare professionals with accurate, data-driven insights to improve care delivery.