It is 2:45 p.m. on a Tuesday in a busy emergency department. Dr. Reyes, a seasoned internist, has just been handed the chart for Mr. Thompson, a 68 year-old man with new-onset atrial fibrillation. He’s also diabetic and living with moderate chronic kidney disease. The ECG confirms the arrhythmia. The labs are back. Heart rate is high but stable. Dr. Reyes pauses at the order screen, hesitating.
Should she anticoagulate now or wait? Is apixaban the best choice for this patient’s renal function, or should she consider something else? Is cardioversion appropriate today? The guidelines are clear when read in isolation, but Mr. Thompson doesn’t fit neatly into the isolated boxes of a guideline section. He fits the real world, with all its clinical nuance and constraints. And now, like so many clinicians, Dr. Reyes faces a decision that is technically supported by guidelines, but practically buried under time pressure, complexity, and fragmented information.
This moment-common, consequential, and deeply human, is where our current systems for guideline dissemination and application often fail. Despite decades of development, most clinical practice guidelines remain difficult to translate into the messy, multifaceted reality of point-of-care decisions.
For those leading the development and dissemination of clinical practice guidelines, this is a familiar tension. You know the effort it takes to synthesize high-quality evidence, balance competing recommendations, and craft text that is both accurate and adaptable. You also know the uphill battle of ensuring that guidance not only reaches clinicians but becomes usable within their workflows. Despite years of digitization efforts, guidelines are still too often static PDFs or fragmented tables embedded in websites, disconnected from clinical systems and removed from the context in which decisions are made.
The Big Tech Proposition and Why It Falls Short
Today, artificial intelligence appears to offer a promising leap forward. And indeed, the major technology players, OpenAI with ChatGPT, Google with Gemini, and others, have captured the imagination of many healthcare institutions by promising natural language interfaces to complex knowledge. Their model is appealing in its simplicity: a large language model, trained broadly on internet-scale data, is coupled with a document retrieval system. When prompted with a question like “Should I anticoagulate this patient with AFib and CKD?”, the system retrieves snippets from guidelines and generates a conversational response.
Yet in clinical practice, this model consistently underdelivers. Studies show that even the most advanced general-purpose LLMs fail to reproduce guideline recommendations reliably. In resuscitation care, for instance, ChatGPT-4 missed 132 of 172 key messages, with errors stemming from superficial or inaccurate reasoning [1]. Similar shortcomings appear in oncology [2] or ophthalmology [3]. These tools can appear fluent and confident, but when pressed on multi-step clinical logic or nuanced contraindications, they fail. And because the model has no memory of the patient beyond what the user types or can retrieve from raw clinical notes, often it cannot reason across time or draw on context not explicitly mentioned in the prompt. The result is a digital assistant that is eloquent, but shallow; accessible, but untrustworthy.
Consider, for instance, how a general-purpose AI might respond to Dr. Reyes’ query about anticoagulation.
Prompt input: “Patient with chronic kidney disease (eGFR 42), history of atrial fibrillation, previously treated with apixaban but discontinued 3 months ago due to GI bleeding. Should I restart anticoagulation?”
Response: “Yes, apixaban is recommended for stroke prevention in patients with atrial fibrillation and a CHADS-VASc score of 2 or more.”
Even though the relevant previous clinical notes were present in the prompt thanks to RAG capabilities, the system does not necessarily structure or prioritize temporal and causal relationships between events. If the system retrieves a recommendation that applies to most patients with atrial fibrillation without accounting for Mr. Thompson’s reduced renal function or fails to recognize that he was previously on apixaban but discontinued it due to gastrointestinal bleeding, information that might reside in a clinical note from three months ago, the result could be a recommendation that is technically plausible, but clinically inappropriate. The system wouldn’t necessarily be wrong, but it wouldn’t be right either. It simply lacks the depth to assess the full patient context, and that margin of ambiguity can matter when clinical decisions carry real-world consequences.
An Integrated, Clinically Grounded Alternative
Other voices in the field have also recognized the limitations of applying general-purpose language models to clinical guideline implementation. For example, researchers evaluating the integration of ChatGPT with COVID-19 outpatient treatment guidelines found that while performance improved with in-context learning and chain-of-thought prompting, the system still produced a moderate rate of hallucinations and failed to consistently reflect the complexity of updated medical criteria [4]. Similarly, efforts to optimize hepatology guideline delivery through structured guideline reformatting, combined with enhanced prompt engineering, revealed that raw LLM outputs showed poor accuracy unless the guideline text was rigorously transformed into a format the model could effectively interpret, raising accuracy from 43% to 99% only after such adaptation [5].
These findings, echoed across domains including ophthalmology [3], thyroid oncology [2], and resuscitation care [1], point to a common conclusion: meaningful use of AI in clinical guideline dissemination demands more than natural language fluency. It requires structure, memory, and alignment with domain logic.
What if we began from this recognition? Rather than treating guidelines and patient data as isolated documents, what if they could be integrated into a shared, structured ecosystem? Imagine a longitudinal representation of the patient, a “patient journey”, constructed dynamically through healthcare-specific NLP systems. This journey would not be a simple list of diagnoses or medications, but a time-aware knowledge graph, enriched with contextual assertions (is a condition active, historical, or ruled out?), temporal markers (when was this lab result flagged as abnormal?), and clinical relationships (was a medication discontinued due to a side effect?). Such a structure could enable an AI system to more fully understand the patient story, even when that story isn’t fully articulated in the clinician’s prompt.
But context alone is not enough. To make these systems truly useful, clinical guidelines themselves must be reimagined. Rather than existing as static prose, they could be represented in structured, computable formats. This notion is not novel. Structured representations have been explored by various groups using ontologies, executable logic, and rule-based modeling. These efforts aim to translate narrative recommendations into logic that systems can evaluate, not just recite [6]. By parsing clinical guidelines into machine-readable representations that reflect decision thresholds, dependencies, and exceptions, it becomes possible to create systems that reason with the text, not just over it.
The combination of a structured patient context and computable guidelines opens the door to a different kind of AI interaction. Not one that simply summarizes retrieved text, but one that reasons across time, adapts to the patient’s unique clinical status, and explains its conclusions. It invites an approach to AI that supports, rather than replaces, the clinician’s reasoning, quietly reducing the cognitive burden of guideline interpretation so that clinicians like Dr. Reyes can focus on the conversation, not the protocol.
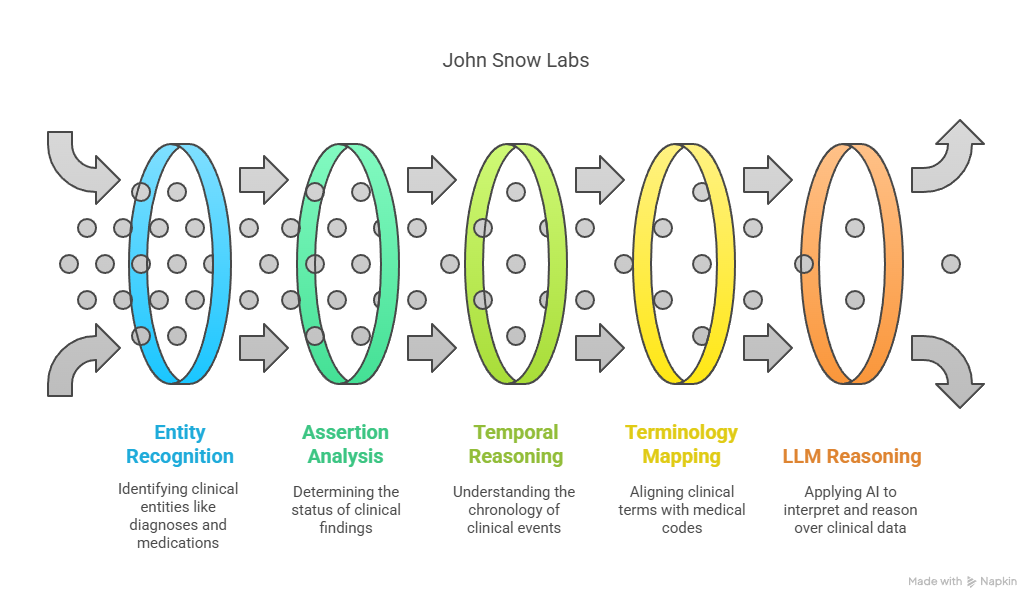
At John Snow Labs, several production-ready components already support the kind of clinically contextualized, guideline-driven AI system envisioned here. The Healthcare NLP library provides more than 200 specialized annotators, including ner_jsl_enriched, a named entity recognition model capable of detecting over 80 clinically relevant entity types, such as diagnoses, medications, lab tests, and procedures. For example, it can accurately extract both “apixaban” and its dosing details from discharge summaries, or identify temporal expressions like “two weeks ago” and resolve them with the DateResolver component.
To understand whether a clinical finding is active, resolved, or hypothetical, the AssertionDLModel and ContextualAssertion modules are used. These allow the system to determine, for example, whether “heart failure” in a clinical note refers to a current diagnosis or a past event no longer affecting treatment decisions.
Temporal reasoning is handled by components such as TemporalRelationExtractor and TimeNormalizer, which can determine that a creatinine result from 10 days ago predates the onset of a new medication, enabling causal or chronological insight in patient timelines.
The ChunkEntityResolver bridges unstructured language with medical terminologies, mapping terms like “blood thinner” to normalized codes in ontologies such as SNOMED CT, ATC, or RxNorm, critical for aligning clinical notes with guideline language.
These NLP outputs can feed into real-time knowledge graphs or patient journey representations, allowing large language models to reason across a patient’s history and current state. Complementing this, a suite of domain-specific Medical LLMs, trained on carefully curated medical datasets, provide state-of-the-art performance on tasks such as clinical question answering, guideline interpretation, and diagnostic triage. For example, the John Snow Labs 14B Medical-Reasoning LLM has demonstrated superior performance in OpenMed benchmark tasks related to treatment planning for multimorbid patients, including cases involving renal and cardiovascular comorbidities.
To illustrate this with a practical example, consider the case of Mr. Thompson. Suppose the source documents include recent clinical notes, a discharge summary, and lab results. The ner_jsl_enriched model identifies mentions of “atrial fibrillation”, “type 2 diabetes”, “chronic kidney disease”, and “apixaban”. It also captures that apixaban was discontinued “three months ago” due to a “gastrointestinal bleed”, a relationship the ContextualAssertion annotator clarifies as being affirmed and relevant to the patient.
The TimeNormalizer and TemporalRelationExtractor determine the correct sequence: the bleeding event occurred after apixaban was initiated and before current admission. The ChunkEntityResolver maps “apixaban” to its corresponding RxNorm and ATC codes, enabling downstream systems to correctly align this treatment history with drug-related guidance in the clinical guideline.
This structured information feeds into a patient-specific knowledge graph, representing Mr. Thompson’s conditions, medication history, lab trends, and temporal context. When a clinical question is posed, such as whether to reinitiate anticoagulation, the system draws not only from retrieved guideline text, but also from this graph, enabling the Medical LLM to reason more accurately.
Original text: “Patient with chronic kidney disease (eGFR 42), history of atrial fibrillation, previously treated with apixaban but discontinued 3 months ago due to GI bleeding.”
Prompt input (JSL-enabled context-aware system): “Should I restart anticoagulation for Mr. Thompson?”
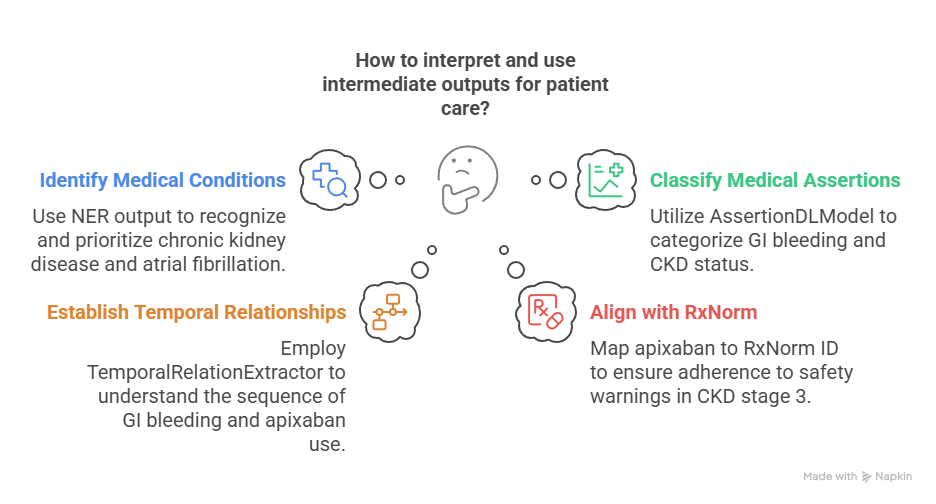
Intermediate outputs:
- ner_jsl_enriched → Identifies “chronic kidney disease”, “eGFR 42”, “atrial fibrillation”, “apixaban”, and “GI bleeding”
- AssertionDLModel → Classifies “GI bleeding” as affirmed, “CKD” as current, and “apixaban” as past treatment
- TemporalRelationExtractor → Links “GI bleeding” as temporally after “apixaban” use
- ChunkEntityResolver → Maps “apixaban” to RxNorm ID 1119620, aligning with DOAC safety warnings in CKD stage 3
This structured knowledge is fed into a longitudinal patient graph. When the LLM receives the query, it leverages the context graph to reason:
- The patient has CHADS-VASc > 2
- Apixaban is contraindicated due to prior GI bleed
- CKD stage 3 modifies DOAC dosing and bleeding risk
Response (JSL-based system): “Given the patient’s CHA₂DS₂-VASc score and prior GI bleeding on apixaban, anticoagulation may still be indicated but with a modified risk-benefit profile. Alternatives such as dose-adjusted apixaban or shared decision-making should be considered per 2020 ESC AFib guidelines, section 4.3.2.”
This example illustrates how the layered reasoning enabled by structured NLP and knowledge graphs offers a fundamentally more reliable and clinically transparent decision pathway.
With these building blocks, it is possible to begin constructing systems that not only retrieve recommendations, but interpret them in light of a patient’s full clinical context—turning clinical guidelines into living tools that accompany, rather than burden, the clinician.
The challenge of bringing guidelines to the bedside is not new. But the tools to meet it have evolved. It is time for guideline developers to move beyond dissemination and toward interaction—for guidance that is not only accessible, but context-aware, computable, and patient-specific. We believe that with the right foundation, the right models, and the right respect for clinical complexity, AI can help us finally close the gap between what we know and what we do.
The next generation of guideline implementation won’t be a better PDF or a more colorful website. It will be a conversation—grounded in structure, enriched by context, and built to serve the clinician, not just impress them.
[1] S. Beck, M. Kuhner, M. Haar, A. Daubmann, M. Semmann, and S. Kluge, “Evaluating the accuracy and reliability of AI chatbots in disseminating the content of current resuscitation guidelines: a comparative analysis between the ERC 2021 guidelines and both ChatGPTs 3.5 and 4,” Scand J Trauma Resusc Emerg Med, vol. 32, no. 1, p. 95, Sep. 2024, doi: 10.1186/s13049-024-01266-2. [2] S. Pandya, T. E. Bresler, T. Wilson, Z. Htway, and M. Fujita, “Decoding the NCCN Guidelines With AI: A Comparative Evaluation of ChatGPT-4.0 and Llama 2 in the Management of Thyroid Carcinoma,” The American SurgeonTM, vol. 91, no. 1, pp. 94–98, Jan. 2025, doi: 10.1177/00031348241269430. [3] M. Balas, E. D. Mandelcorn, P. Yan, E. B. Ing, S. A. Crawford, and P. Arjmand, “ChatGPT and retinal disease: a cross-sectional study on AI comprehension of clinical guidelines,” Canadian Journal of Ophthalmology, vol. 60, no. 1, pp. e117–e123, Feb. 2025, doi: 10.1016/j.jcjo.2024.06.001. [4] Y. Wang, S. Visweswaran, S. Kapoor, S. Kooragayalu, and X. Wu, “ChatGPT-CARE: a Superior Decision Support Tool Enhancing ChatGPT with Clinical Practice Guidelines,” Aug. 13, 2023. doi: 10.1101/2023.08.09.23293890. [5] S. Kresevic, M. Giuffrè, M. Ajcevic, A. Accardo, L. S. Crocè, and D. L. Shung, “Optimization of hepatological clinical guidelines interpretation by large language models: a retrieval augmented generation-based framework,” npj Digit. Med., vol. 7, no. 1, p. 102, Apr. 2024, doi: 10.1038/s41746-024-01091-y. [6] V. Zamborlini et al., “Analyzing interactions on combining multiple clinical guidelines,” Artificial Intelligence in Medicine, vol. 81, pp. 78–93, Sep. 2017, doi: 10.1016/j.artmed.2017.03.012.