Converting free-text medical descriptions into structured ontology codes with validation
Human phenotypes, observable traits and clinical abnormalities like “short stature” or “muscle weakness” are crucial in diagnosing diseases, especially in rare and genetic conditions. However, these phenotypes are often buried in unstructured clinical text. To address this, we built an NLP pipeline using John Snow Labs’ Healthcare NLP & LLM library that automatically extracts phenotype mentions, determines their assertion status (e.g., present, absent, etc.), and maps them to standardized Human Phenotype Ontology (HPO) codes, which provides a structured vocabulary used worldwide in genomics and precision medicine. This pipeline enables scalable, accurate phenotypic data extraction for research and clinical applications.
Introduction
In modern medicine, the ability to capture and analyze patient phenotypes, the observable traits and clinical signs associated with disease is critical for accurate diagnosis, disease understanding, and personalized care. Human phenotypes such as ataxia, muscle weakness, or global developmental delayoften hold the key to unlocking complex clinical patterns, especially in rare or genetic disorders. Yet, despite their importance, these phenotypic descriptions are typically scattered throughout unstructured clinical notes, case reports, or biomedical literature, making them difficult to access or analyze systematically.
To address this, the Human Phenotype Ontology (HPO) was developed as a standardized vocabulary for phenotypic abnormalities encountered in human disease. It includes over 18,000 terms arranged in a hierarchical structure and is used globally in genomics, rare disease research, and clinical diagnostics to ensure consistent annotation of patient data. However, leveraging HPO in real-world settings requires the ability to extract relevant terms from raw text and accurately link them to their ontology entries.
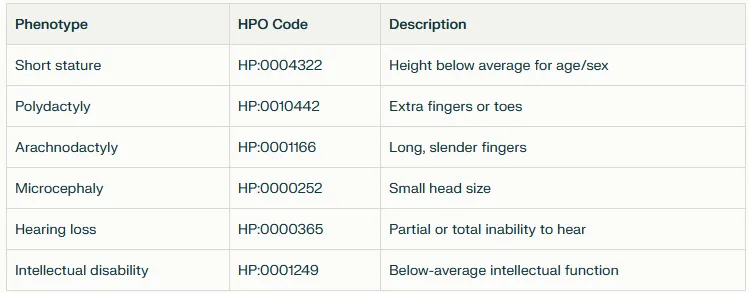
Here are a few common human phenotypes with their corresponding HPO codes:
This is where Natural Language Processing (NLP) plays a transformative role. In this blog post, we introduce an NLP pipeline that automates the process of extracting human phenotype mentions from unstructured text, determines their assertion status (e.g., whether the phenotype is present, absent, suspected, or conditional), and then maps each entity to the correct HPO code. This approach not only streamlines information extraction but also enables downstream applications such as patient stratification, cohort selection, and computational phenotyping.
Whether you’re a clinical researcher, data scientist, or bioinformatician, this pipeline provides a scalable and accurate way to convert rich but messy clinical narratives into structured, machine-readable data — unlocking the true potential of phenotypic information for precision medicine.
You can use this notebook to explore the process of extracting human phenotypes and their corresponding HPO codes.
Understanding Human Phenotypes and Their Clinical Prevalence
Human phenotypes are the observable characteristics, signs, and symptoms resulting from the interaction between an individual’s genes and the environment. In clinical practice, these can range from specific findings like ataxia or macrocephaly to broader features such as global developmental delayor hearing impairment. These phenotypic features are essential for diagnosis, especially in rare and genetic diseases, where phenotype-driven approaches are often the first step in identifying candidate genes or conditions.
Phenotypic data appears in over 80% of electronic health record (EHR) narratives, making it one of the most abundant and clinically relevant types of information in unstructured clinical text according to a major research effort. In rare disease diagnostics, phenotype descriptions are even more critical: a 2020 study from the NIH Undiagnosed Diseases Program found that 97% of diagnostic decisions were informed by detailed phenotypic profiles. The Human Phenotype Ontology (HPO), developed to formalize and structure these phenotype terms, is a cornerstone of modern phenomics. It currently contains over 18,000 standardized terms and is used by resources like OMIM, ClinVar, and Orphanet to annotate gene-disease relationships.
In genomics workflows, HPO terms are widely used to prioritize variants during exome and genome sequencing analyses. Studies have shown that phenotype-driven algorithms using HPO can significantly improve diagnostic yield by narrowing down candidate genes based on phenotypic similarity scores. Despite their importance, phenotypic mentions are often locked in unstructured text, making them inaccessible to computational systems unless extracted and normalized using NLP tools.
By automatically identifying phenotype mentions, classifying their assertion status (e.g., present, absent, etc.), and linking them to HPO codes, NLP solutions can transform unstructured clinical narratives into rich, structured datasets that fuel diagnostics, cohort selection, and translational research.
Frequency of Human Phenotype–Defined Diseases
Understanding the frequency of diseases associated with human phenotypes provides valuable context for why automated extraction and normalization of phenotypic information is essential. While some conditions are relatively common and well-documented, others are rare yet critically important for diagnosis and treatment planning. Below are examples of both common and rare diseases, each with characteristic phenotypic patterns that highlight the diverse landscape in which phenotype-driven NLP systems can make a significant impact.
- Cystic Fibrosis, a recessive genetic disease, affects ~30,000 people in the U.S., with a carrier frequency of approximately 1 in 25 among European Americans.
- Celiac Disease, a common autoimmune condition, has a prevalence of about 1 in 100–170 globally, though up to 85% of cases may be undiagnosed.
- Osteogenesis Imperfecta occurs in roughly 1 in 20,000 live births in the U.S., totaling 20,000–50,000 affected individuals.
- Friedreich’s Ataxia, the most common inherited ataxia in European populations, shows a prevalence of ~1 in 40,000, with carrier rates around 1 in 100.
- Huntington’s Disease, a late-onset neurodegenerative disorder, has an average prevalence of 5–10 per 100,000 worldwide, but up to ~12 per 100,000 in northern Europe.
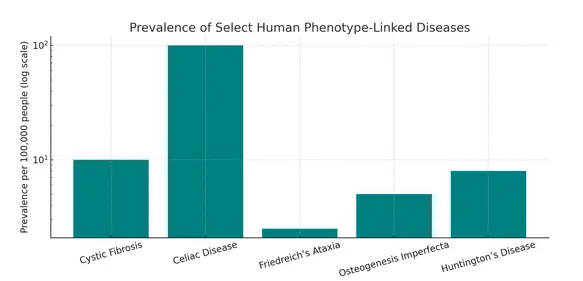
Prevalence of Selected Human Phenotype–Linked Diseases (on a log scale).
Why These Numbers Matter
- High prevalence of certain genetic diseases (e.g., 1 in 40,000 carriers) emphasizes that impactful phenotypic patterns recur often enough for efficient NLP extraction.
- Rich phenotype-disease associations (e.g., average 5–10 HPO terms per disease) underscore the complexity of accurate mapping and the necessity of assertion detection.
- Symptom frequency distributions indicate that many critical disease traits are frequent or very frequent, making them amenities for pattern recognition yet rarer symptoms still carry high diagnostic value.
Extracting Entities from Clinical/Medical Text
Pretrained pipelines in the Healthcare NLP library enable the systematic extraction and organization of hidden information from text, transforming it into structured datasets that support advanced analysis.
In our case, we use one line of code to process the pretrained pipeline (hpo_mapper_pipeline_v2), which is specifically trained to extract human phenotype entities and assign the related HPO codes:
pipeline = PretrainedPipeline("hpo_mapper_pipeline_v2", "en", "clinical/models")
result = pipeline.fullAnnotate(text)
Extracting entities in a structured format enhances usability and integration by allowing efficient retrieval and in-depth analysis of patient information. It promotes consistency and standardization, which supports advanced analytics and more accurate decision-making. By converting raw text into actionable insights, this approach contributes to improved patient care, more effective research, and better-informed public health strategies.
In our case, the extracted NER and assertion results for human phenotypes were returned as a DataFrame, making them easy to explore, manipulate, and integrate into downstream pipelines.
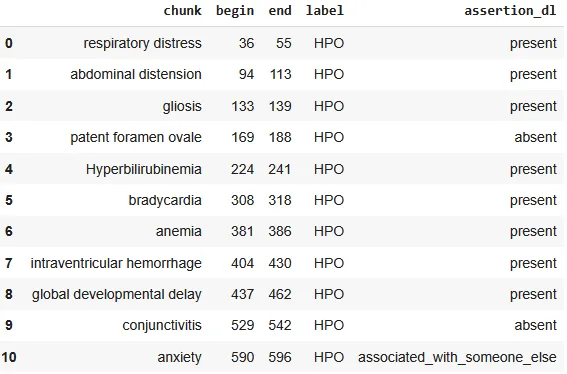
Dataframe providing the chunks, assigned labels and assertion status.
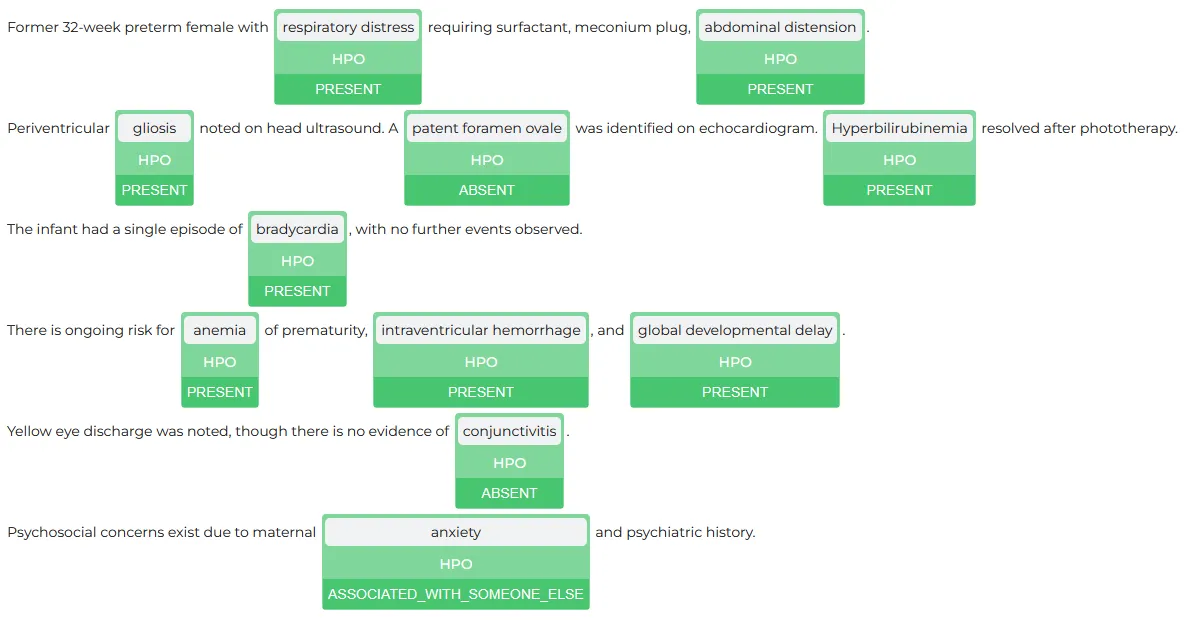
The ability to quickly visualize the entities is a very useful feature for examining the generated results. Spark NLP Display is an open-source Python library for visualizing the extracted and labeled entities. NerVisualizer highlights the extracted named entities and also displays their labels as decorations on top of the analyzed text.
AssertionVisualizer is a special type of NerVisualizer that also displays on top of the labeled entities the assertion status that was inferred by a Healthcare NLP model.
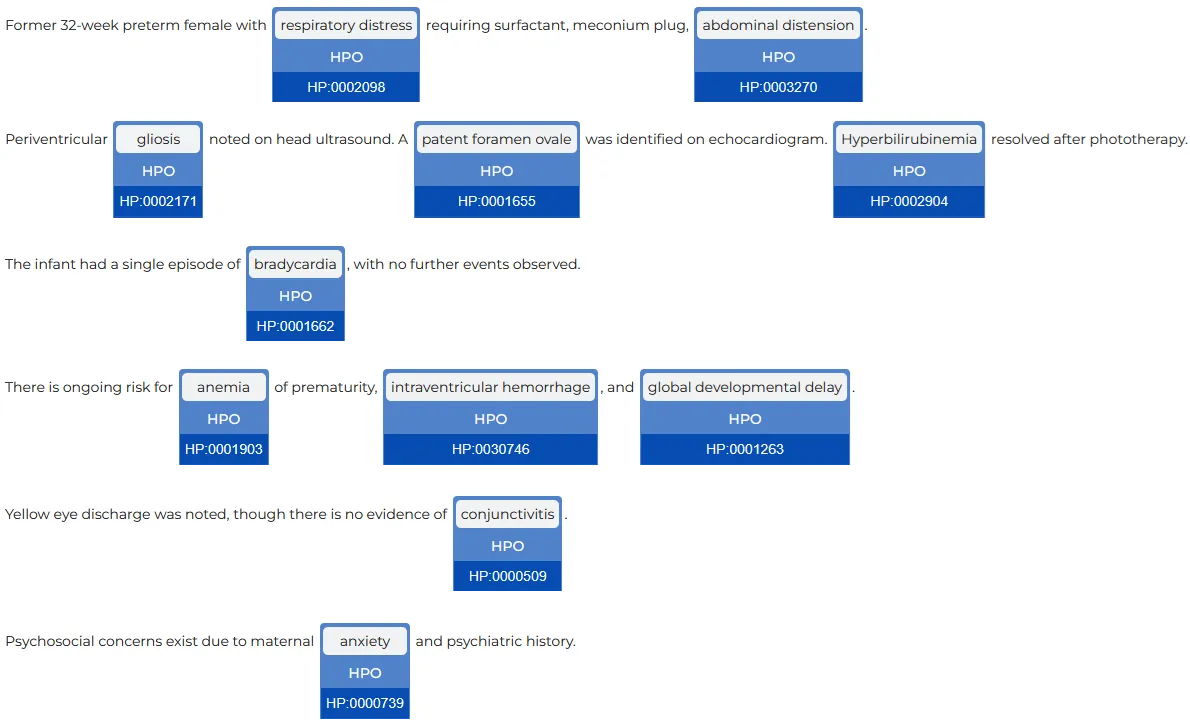
The HPO codes are added as a new column to the previous dataframe. We can post-process the dataframe and filter for Present only.
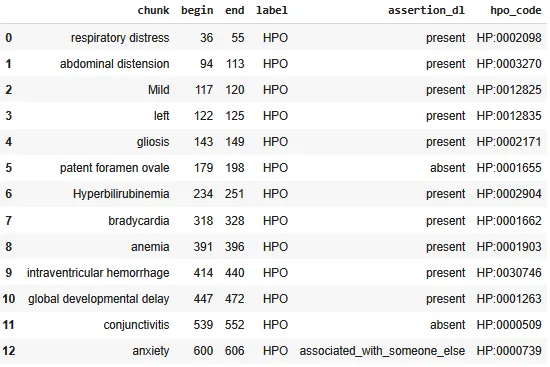
Dataframe providing the chunks, assigned labels and assertion status with the HPO codes.
EntityResolverVisualizer will automatically display on top of the NER label the standard code (ICD10 CM, PCS, CPT, HPO etc.) that corresponds to that entity.
By combining NER, assertion status detection, and HPO code resolution, we’ve built an NLP pipeline capable of transforming unstructured clinical text into structured, ontology-linked phenotype data. This approach not only streamlines phenotypic data extraction but also enhances consistency, searchability, and downstream analytics. With outputs returned in a clean, tabular format, such as a DataFrame, this solution is well-suited for integration into clinical research, rare disease diagnosis, and precision medicine workflows. As phenotype-driven methods become increasingly important, automating this process is a crucial step toward scalable and intelligent healthcare data analysis.
You can view the details of Human Phenotype Extraction and HPO Code Mapping, check out the official Healthcare NLP notebook in John Snow Labs Github repository.
Conclusion
The application of John Snow Labs’ Healthcare NLP and LLM library to genetic and phenotype text analysis represents a major breakthrough in the field of medical research.
In this blog post, we demonstrated how an NLP pipeline can effectively bridge the gap between free-text clinical text and structured phenotype data. By integrating NER, assertion status detection, and HPO code resolution, our solution captures not only the mention of human phenotypes but also their contextual meaning whether a condition is present, absent, hypothetical etc.and links them to standardized ontology terms. This level of semantic precision is essential for enabling interoperability, improving data quality, and ensuring that phenotype information can be reliably used across clinical and research systems.
The resulting structured outputs, organized into a clear and queryable DataFrame format, are ideal for downstream tasks such as patient stratification, cohort discovery, variant prioritization, and population health studies. As the healthcare landscape continues to shift towards data-driven decision-making, the importance of robust and scalable NLP solutions will only continue to grow. By automating the process of phenotype extraction and standardization, we can empower researchers, clinicians, and analysts to focus on high-value tasks, driving meaningful insights and breakthroughs in patient care. With our solution, we aim to contribute to the development of more intelligent, efficient, and effective healthcare data analysis, ultimately improving patient outcomes and advancing the field of precision medicine.
John Snow Labs and Healthcare NLP
John Snow Labs, offers a powerful NLP & LLM library tailored for healthcare, empowering professionals to extract actionable insights from medical text. Utilizing advanced AI techniques like Named Entity Recognition (NER), assertion status detection, relation extraction, Question-Answering, and summarizing, this library helps uncover vital genetics information for more accurate diagnosis, treatment, and prevention.
Healthcare NLP
The Healthcare Library is a powerful component of John Snow Labs’ Healthcare NLP platform, designed to facilitate NLP tasks within the healthcare domain. This library provides over 2,700 pre-trained models and pipelines tailored for medical data, enabling accurate information extraction, NER for clinical and medical concepts, and text analysis capabilities. Regularly updated and built with cutting-edge algorithms, the Healthcare library aims to streamline information processing and empower healthcare professionals with deeper insights from unstructured medical data sources, such as electronic health records, clinical notes, and biomedical literature.
John Snow Labs has created custom large language models (LLMs) tailored for diverse healthcare use cases. These models come in different sizes and quantization levels, designed to handle tasks such as summarizing medical notes, answering questions, performing retrieval-augmented generation (RAG), named entity recognition and facilitating healthcare-related chats.
John Snow Labs’ GitHub repository serves as a collaborative platform where users can access open-source resources, including code samples, tutorials, and projects, to further enhance their understanding and utilization of Healthcare NLP and related tools.
John Snow Labs also offers periodic certification training to help users gain expertise in utilizing the Healthcare Library and other components of their NLP platform.
John Snow Labs’ demo page provides a user-friendly interface for exploring the capabilities of the library, allowing users to interactively test and visualize various functionalities and models, facilitating a deeper understanding of how these tools can be applied to real-world scenarios in healthcare and other domains.