TL; DR
This post presents a focused update on large-scale clinical de-identification benchmarks, emphasizing pipeline design, execution strategy, and infrastructure-aware performance. Rather than treating accuracy as an isolated metric, we analyze how different pipeline architectures — rule-augmented NER, hybrid NER + zero-shot, and zero-shot–centric approaches — behave under realistic Google Colab and Databricks–AWS deployments.
Clinical De-identification Pipelines
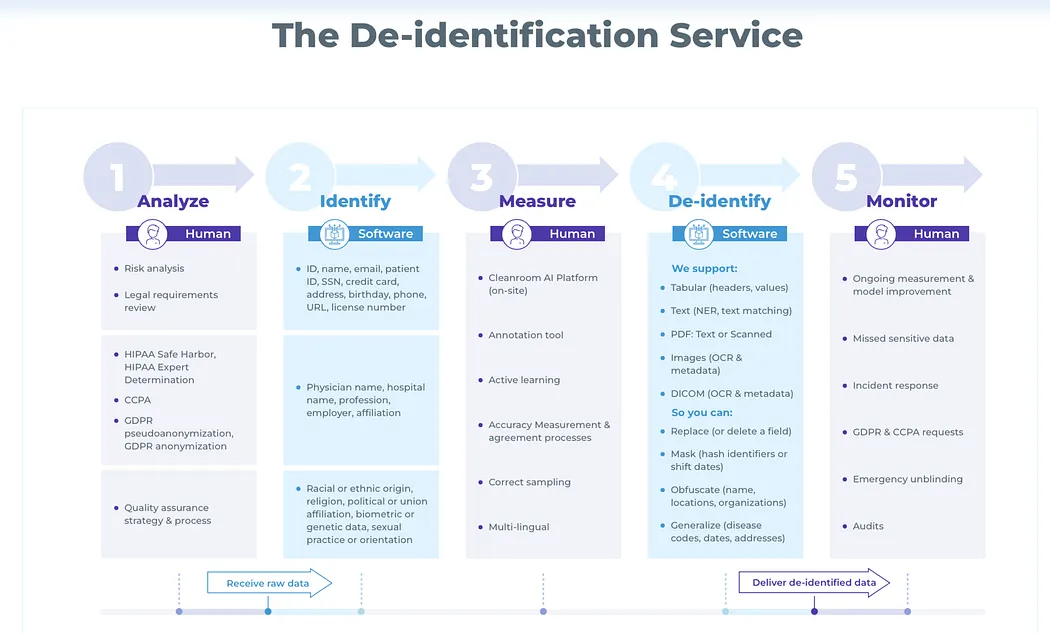
Clinical de-identification pipelines in Spark NLP for Healthcare are designed as composable systems, not monolithic models. Each pipeline reflects an explicit trade-off between precision guarantees, recall robustness, runtime cost, and operational complexity.
Before reviewing benchmark results, it is important to clarify why these pipelines differ and what problem each is intended to solve.
Rule-Augmented Clinical NER Pipelines
Design objective: Deterministic precision for regulated PHI categories
These pipelines combine clinical NER models with a comprehensive set of rule-based annotators for structured identifiers such as dates, phone numbers, IDs, emails, and numeric patterns.

Characteristics
- High precision and predictable behavior
- Strong coverage for explicitly defined PHI types
- Minimal variance across datasets
Trade-offs
- Increased latency as rule count and merge stages grow
- Limited generalization to unseen or context-dependent PHI
Such pipelines are commonly used in compliance-critical and audit-heavy environments, where deterministic behavior is prioritized over maximal recall.
Hybrid NER + Zero-shot Pipelines
Design objective: Balanced recall and robustness with controlled complexity
Hybrid pipelines reduce the number of NER stages and introduce zero-shot chunking models to capture PHI patterns that are difficult to enumerate or label exhaustively.
Characteristics
- Improved recall on ambiguous or long-span entities
- Better generalization across institutions and note styles
- Reduced architectural depth compared to full multi-stage pipelines
Trade-offs
- Higher per-stage computational cost
- Greater sensitivity to batch size and hardware configuration
Hybrid architectures are typically preferred for production systems processing heterogeneous clinical corpora.
Zero-shot De-identification Pipelines
Design objective: Generalization and rapid domain transfer
Zero-shot pipelines rely primarily on pretrained language models capable of span-level PHI detection without task-specific fine-tuning, optionally constrained by lightweight rule-based filters.
Characteristics
- Strong performance on unseen PHI categories
- Reduced dependency on labeled clinical data
- Simplified pipeline structure
Trade-offs
- GPU dependency for production-scale throughput
- Higher inference cost compared to NER
As discussed in John Snow Labs technical content and product documentation, zero-shot clinical NER models reduce annotation requirements and prioritize inference-time efficiency and infrastructure tuning for real-world deployments.
Datasets Used in This Study
Four complementary datasets are used to evaluate clinical de-identification pipelines from different perspectives. Dataset 1 and Dataset 2 focus on accuracy, combining expert-annotated ground truth with a larger surrogate dataset for robustness testing. Dataset 3 is designed for speed and throughput evaluation, using long clinical documents to measure end-to-end pipeline performance. Dataset 4 is used for cloud-scale speed comparisons on Databricks–AWS, enabling performance analysis across different de-identification pipelines and pretrained zero-shot NER configurations.
Dataset 1: Paper Dataset — Expert-Annotated
The first dataset used in this study, referred to as the paper dataset, is an expert-annotated clinical benchmark designed to evaluate the accurate identification of patient identifiers in clinical narratives. The dataset was created using John Snow Labs’ Annotation Lab, following a multi-stage annotation workflow. This process began with a deep learning–based pre-annotation step to extract candidate entities, followed by multiple rounds of expert review and refinement guided by a dynamic annotation guideline. This iterative approach ensured high annotation quality, consistency, and adaptability throughout the evaluation process.
The paper dataset consists of 48 clinical notes and was specifically curated to support the evaluation of healthcare de-identification systems. Expert annotators focused on six core categories of Protected Health Information (PHI): NAME, DATE, LOCATION, AGE, CONTACT, and IDNUM. These entity types were selected due to their frequent occurrence in clinical text and their importance under regulatory frameworks such as the Health Insurance Portability and Accountability Act (HIPAA) and the General Data Protection Regulation (GDPR).
Direct identifiers (e.g., names, contact details, and identification numbers) pose a high risk of re-identification if not properly removed, while quasi-identifiers such as dates, locations, and age can enable indirect re-identification when combined with other information. By centering the benchmark on these categories, the paper dataset reflects real-world de-identification challenges commonly encountered in clinical settings.
To support transparency and reproducibility, the paper dataset has been made publicly available via a dedicated repository.
Dataset 2: Curated Surrogate Clinical Dataset
The second dataset is a privately curated surrogate clinical dataset developed to complement the paper dataset and to enable large-scale, realistic evaluation scenarios. The dataset consists of 238 clinical documents and does not contain real patient records; instead, it is composed of synthetic and surrogate clinical text designed to closely mirror the linguistic structure and information density of real clinical documentation.
The dataset was curated in-house with a focus on increasing PHI diversity and density, enabling stress testing of de-identification systems under more challenging and realistic conditions. It was primarily used to evaluate system robustness, scalability, error patterns, and edge-case behavior beyond what is feasible with smaller public datasets.
Annotations cover key PHI categories commonly encountered in clinical workflows, including NAME, DATE, LOCATION, AGE, CONTACT, and ID.
Dataset 3: Document-Level Clinical De-Identification Context Dataset
Dataset 3 is a document-level clinical dataset specifically designed to benchmark the throughput and latency performance of clinical de-identification pipelines under realistic long-context conditions. It consists of 1,000 clinical documents, each with an average length of 503 tokens, approximating the size and structural complexity of real-world clinical reports.
Dataset 4: Large-Scale Aggregated Clinical NER and De-Identification Dataset
This dataset is a large-scale aggregated clinical dataset constructed to support extensive evaluation and stress testing of medical named entity recognition (NER) and de-identification systems. It was created by merging heterogeneous clinical data sources, including 1,100 distinct clinical notes, over 360,000 medical NER reports, and more than 110,000 masked PII clinical notes. This consolidation strategy was designed to capture a wide range of clinical writing styles, documentation formats, and entity distributions commonly observed in real-world healthcare data.
The resulting dataset comprises a total of 478,527 rows, with an average text length of 152.71 characters per record. In aggregate, the dataset contains approximately 11.7 million tokens, with an average of 24.46 tokens per row, making it well-suited for large-scale model training, benchmarking, and throughput evaluation.
It emphasizes both medical entity diversity and privacy-aware text representations. The inclusion of masked PII notes enables controlled evaluation of de-identification robustness, while the large volume of NER medical reports supports fine-grained analysis of token-level prediction accuracy, boundary detection, and error propagation across varied clinical contexts. By combining raw clinical narratives with pre-processed and privacy-preserving variants, the dataset facilitates comprehensive assessment across multiple stages of healthcare NLP pipelines.
Clinical De-identification — Most Up-to-Date Pipelines
John Snow Labs’ Healthcare NLP library provides a large collection of pretrained models and pipelines designed specifically for clinical and biomedical text processing. Within this ecosystem, clinical de-identification pipelines represent the most mature and operationally validated solutions for automated PHI detection and removal, combining deterministic rules, pretrained NER models, and zero-shot models.
Rather than relying on a single modeling approach, these pipelines are designed to achieve regulatory-grade accuracy under real-world constraints, where throughput, infrastructure availability, and cost efficiency are as critical as raw detection performance.
Before presenting benchmark results, we outline the experimental setup used to evaluate these pipelines in a controlled and reproducible manner.
Dataset and Experimental Setup
Datasets Used
- Dataset 1: Paper Dataset — Expert-Annotated
- Dataset 2: Curated Surrogate Clinical Dataset
- Dataset 3: Document-Level Clinical De-Identification Context Dataset
Key dataset properties:
- Identical documents and token distributions were used across all pipeline configurations
- No preprocessing or normalization steps were introduced that could bias runtime or accuracy
- The dataset was used exclusively for end-to-end clinical de-identification benchmarking, with a focus on token-level PHI detection
This controlled setup ensures that observed differences are attributable to pipeline architecture and execution strategy, rather than data variability.
Compute Environments
To reflect common deployment scenarios, two execution environments were evaluated.
GPU-Based Spark Environment
- Hardware: NVIDIA A100 GPU
- Execution framework: Apache Spark
- Configuration: 48 Spark partitions
This setup represents a production-grade GPU environment optimized for high-throughput clinical NLP inference.
CPU-Based Spark Environment
- Hardware: Google Colab High-RAM CPU
- Execution framework: Apache Spark
- Configuration: 32 Spark partitions
This environment models a CPU-only batch processing scenario, commonly used for offline processing or cost-constrained deployments.
Benchmark Procedure
Each pipeline was evaluated using an identical, step-by-step benchmark process.
Input preparation
- All 1,000 clinical documents were ingested without modification
- Token counts and sentence boundaries were preserved to avoid preprocessing bias
Pipeline execution
Each clinical de-identification pipeline was executed independently on:
- the GPU-based Spark cluster, and
- the CPU-based Spark cluster
Used Pipelines
- clinical_deidentification_docwise_benchmark_optimized
- clinical_deidentification_docwise_benchmark_medium
- clinical_deidentification_docwise_benchmark_medium_v2
- clinical_deidentification_docwise_zeroshot_medium
- clinical_deidentification_docwise_zeroshot_medium (Single Stage)
- clinical_deidentification_docwise_benchmark_large
- clinical_deidentification_docwise_benchmark_large_v2
- clinical_deidentification_docwise_zeroshot_large
- clinical_deidentification_docwise_zeroshot_large (Single Stage)
Evaluation metrics
Token-level precision, recall, and F1-score were computed using:
- paper-based annotations as the primary reference
- surrogate annotations as a secondary evaluation signal
Controlled comparison
- Dataset size, token distribution, and evaluation methodology were held constant across all pipelines
- No caching or intermediate reuse was applied
This methodology isolates performance–accuracy trade-offs under identical experimental conditions.
Benchmark Results
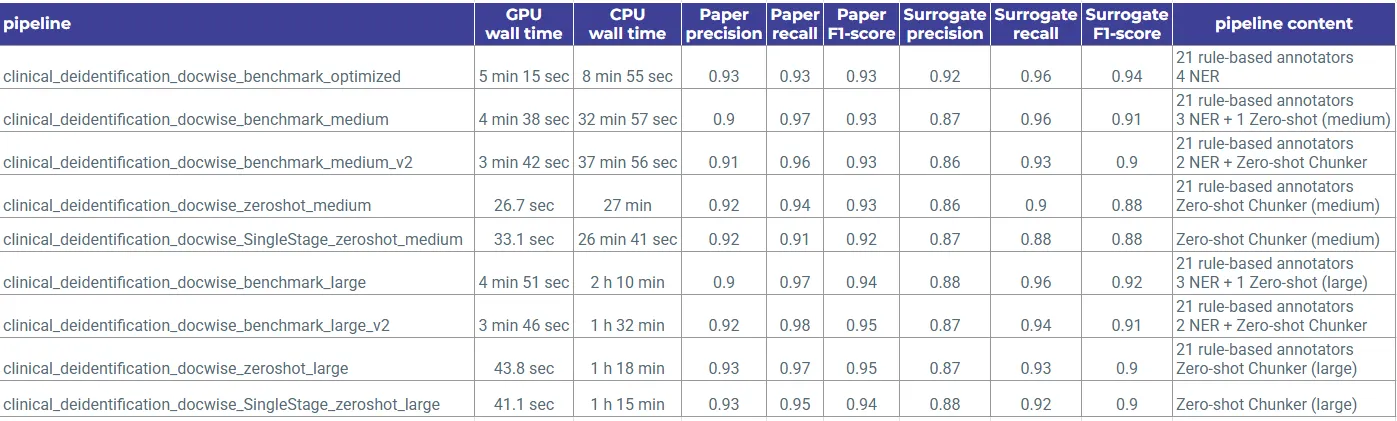
The benchmark results highlight clear and repeatable differences across pipeline architectures.
Zero-shot models perform span-level PHI detection without task-specific fine-tuning, relying on general language understanding rather than label alignment.
Two zero-shot model scales were evaluated:
Medium zero-shot models
Faster inference with slightly lower recall, suitable for throughput-sensitive deployments
Large zero-shot models
Higher recall and robustness, particularly for ambiguous or long-span PHI, at increased computational cost
Model Comparison by Pipeline Type
To enable a structured comparison, pipelines were grouped by architectural complexity.
- Multi-stage pipelines
Combine rule-based annotators, multiple clinical NER models, and a zero-shot chunker
Prioritize recall and coverage across diverse PHI categories - Hybrid pipelines
Use fewer NER models combined with a zero-shot chunker
Reduce architectural depth while preserving robustness - Single-stage pipelines
Rely primarily on a zero-shot chunker
Optionally apply lightweight rule-based filtering
Summary
Across all configurations, the evaluated pipelines leverage deterministic rules, pretrained NER models, and zero-shot language models in varying combinations.
This design enables a controlled comparison of:
- model scale
- architectural complexity
- runtime behavior
- accuracy trade-offs
Together, these benchmarks provide a clear foundation for understanding how modern clinical de-identification systems behave under realistic infrastructure and execution constraints.
De-identification Pipelines Speed Comparison on Databricks–AWS
With pipeline design clarified, we now examine how architectural choices interact with infrastructure at scale.
Experimental Context
Dataset Used
- Dataset 4: Large-Scale Aggregated Clinical NER and De-Identification Dataset (~478K clinical documents , ~11.7M tokens)
Platform
- Databricks on AWS
Configurations
Databricks-AWS Config with CPU/GPU Options:
- (CPU) Worker Type: m5d.2xlarge 32 GB Memory, 8 Cores, 8 Workers
- (GPU) Worker Type: g4dn.2xlarge[T4] 32 GB Memory, 1 GPU, 8 Workers
Pipelines evaluated
clinical_deidentification_docwise_benchmark_large
clinical_deidentification_docwise_benchmark_optimized
clinical_deidentification_docwise_benchmark_medium
Benchmark Results

CPU Runtime Comparison of All Three Pipelines

CPU & GPU Runtime Comparison of Medium Pipeline
CPU & GPU Runtime Comparison of Medium Pipeline
- These benchmark results demonstrate the computational impact of pipeline design and infrastructure choice on large-scale clinical deidentification workloads executed on Databricks-AWS. Using a substantial and diverse clinical corpus (~478K documents, ~11.7M tokens), the results highlight clear performance trade-offs.
- Pipeline complexity is the primary driver of runtime. The Large pipeline, with the most extensive model stack, exhibits the highest execution time (9.4 hours on CPU). The Medium pipeline achieves a notable reduction (~3.1 hours on CPU), while the Optimized pipeline delivers a step-change improvement, completing in ~26 minutes on CPU due to architectural simplifications and reduced model overhead.
- GPU acceleration further enhances performance for the Medium pipeline, reducing execution time from ~3.1 hours (CPU) to ~1.0 hour (GPU), even with a smaller batch size. This indicates that GPU utilization effectively mitigates inference bottlenecks in moderately complex NLP pipelines.
Overall, the findings emphasize that pipeline optimization yields greater performance gains than hardware scaling alone, while GPU resources provide additional, complementary speedups when applied to appropriately balanced pipeline configurations.
Pretrained Zero-Shot NER De-identification Subentity Speed Comparison on Databricks–AWS
Zero-shot Named Entity Recognition (NER) enables the identification of entities in text with minimal effort. By leveraging pre-trained language models and contextual understanding, zero-shot NER extends entity recognition capabilities to new domains and languages.
This experiment compares the Pretrained Zero-shot NER runtime for CPU and GPU clusters on Databricks-AWS environment.
Experimental Context
Dataset Used
- Dataset 4: Large-Scale Aggregated Clinical NER and De-Identification Dataset (~478K clinical documents , ~11.7M tokens)
Platform
Databricks-AWS Config with CPU/GPU Options
- (CPU) Worker Type: m5d.2xlarge 32 GB Memory, 8 Cores, 8 Workers
- (GPU) Worker Type: g4dn.2xlarge[T4] 32 GB Memory, 1 GPU, 8 Workers
Pipelines evaluated
Benchmark Results

Zero-shot Medium Model CPU & GPU Runtime Comparison
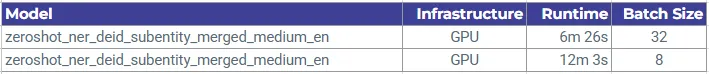
Zero-shot Medium Model Batch Size Comparison via GPU Cluster

Zero-shot Medium & Large Models GPU Runtime Comparison
Zero-shot Medium & Large Models GPU Runtime Comparison
These results highlight the significant impact of GPU acceleration, batch size tuning, and model scale on deidentification pipeline runtime.
- CPU vs GPU (Medium model):
GPU execution provides an orders-of-magnitude speedup, reducing runtime from ~2.8 hours on CPU to ~6.5 minutes on GPU at the same batch size. This clearly indicates that the Medium model is compute-bound on CPU and highly optimized for GPU inference. - GPU batch size comparison (Medium model):
Increasing the batch size from 8 to 32 nearly halves the runtime (from ~12.0 minutes to ~6.4 minutes). This demonstrates that throughput scales efficiently with larger batches on GPU, provided memory constraints are respected. - Medium vs Large model on GPU:
At the same batch size (8), the Large model requires ~30.4 minutes, compared to ~12.0 minutes for the Medium model. This reflects the expected cost of increased model complexity, confirming a direct trade-off between model capacity and inference speed.
Overall, the findings show that GPU usage is essential for production-scale runs, batch size optimization is critical for maximizing GPU efficiency, and model size should be selected based on the required balance between accuracy and runtime performance.
Final Takeaway
Clinical de-identification at scale is fundamentally a systems engineering problem. Accurate models are necessary, but insufficient on their own. Real-world performance is determined by how model capacity, pipeline architecture, Spark execution strategy, and infrastructure choice are aligned.
Several key conclusions can be drawn from the benchmarks:
- Pipeline complexity is the dominant factor in runtime. Each additional NER model, zero-shot component, or chunk merging stage introduces measurable latency. Optimized pipelines consistently outperform larger architectures, even on the same hardware.
- GPU acceleration provides significant but selective benefits. GPUs deliver major speedups for zero-shot and medium-complexity pipelines, but do not compensate for overly complex multi-stage designs.
- Partitioning and batch size tuning are critical. Both under- and over-parallelization degrade performance. Optimal configurations exist in narrow ranges and must be tuned per infrastructure.
- Zero-shot models trade speed for generalization. They excel in robustness and unseen PHI detection but require GPU execution and careful batching to remain production-viable.
References and Further Reading (Selected Technical Resources)
The following resources provide additional technical background on clinical de-identification pipelines, zero-shot named entity recognition, and infrastructure-aware NLP system design. These materials are intended to support the architectural concepts and benchmarking methodology discussed in this post.
- Simpler & More Accurate Deidentification in Spark NLP for Healthcare
Description of de-identification models and pipeline components in Spark NLP for Healthcare. - Effortless PHI De-Identification: Running De-identification and Obfuscation in Healthcare NLP
Overview of de-identification annotators and multi-mode masking/obfuscation strategies. - Pretrained Zero-Shot Models for Clinical Entity Recognition
Discussion of zero-shot NER models for clinical text, focusing on reduced annotation requirements, domain generalization, and inference-time considerations. - Advanced NLP Techniques: Identifying Named Entities in Medical Text with Zero-Shot Learning
Technical description of span-level entity detection using zero-shot learning and its role in hybrid clinical NLP pipelines. - Spark NLP Architecture and Performance Considerations
Background on Spark-based NLP execution strategies, including partitioning, batch size tuning, and orchestration overhead in distributed clinical NLP systems. - Deidentification Benchmarks
Benchmark results illustrating the impact of pipeline complexity and infrastructure configuration on large-scale clinical de-identification workloads.
Readers seeking deeper methodological detail are encouraged to consult the peer-reviewed papers , related academic publications, and technical blogs by the John Snow Labs research team.





























