The last blog (A Primer to EBM – Part [A]) introduced in brief the well-known healthcare research databases, their structure, and the steps followed to determine the best and most recent clinical decision.
This blog will extend further the structure of the PubMed database and how we can work with it to extract the best clinical guidelines or the best clinical decision.
UNDERSTANDING MESH
The National Library of Medicine’s Medical Subject Headings (MeSH)
It is a well-organized and indexed nomenclature for medical terms supplied by the US National Library of Medicine. MeSH indexing can be considered a summary for the medical literature where the data quality is guaranteed and well-reviewed.
The MeSH database is available for download here.
The recent ASCII MeSH download file is: d2019.bin
Due to its hierarchies and branching structure, most researchers describe MeSH as a tree structure, but Jules J. Berman thinks that this is not an accurate description as a single entry may be assigned multiple Mesh Numbers (MN). This means that different branches and nodes can be interconnected together.
The record for a MeSH term contains:
– A definition of the term.
– Associated subheadings.
– A list of entry terms.
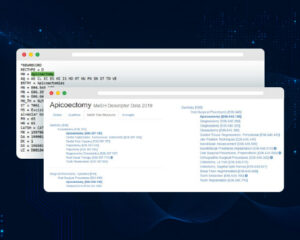
The following is a sample of a MeSH record:

Figure 1: How does a MeSH record look like?
A MeSH record can be analyzed as follows:
– MeSH Term “Apicoectomy” is assigned 3 MeSH Numbers:
MN = E04.545.100
MN = E06.397.102
MN = E06.645.100
By removing the last set of numbers after the last decimal, we can obtain the Mesh Number of the “Parent Term”.
This can be explained as follows:
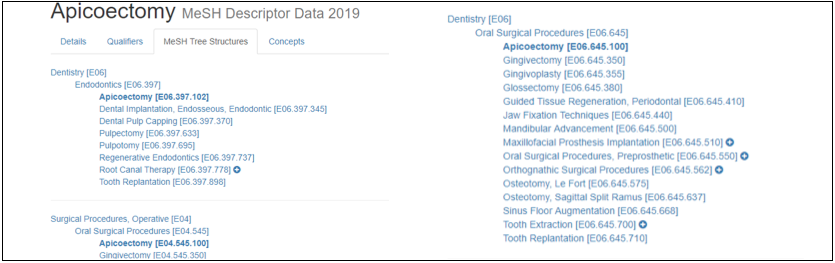
Figure 2: MeSH Parent Term
Note that, each “Parent Term” can be assigned multiple Mesh Numbers, where each one can yield a multibranch hierarchy and therefore Jules J. Berman considered it a non-tree structure as explained before.[1]
Many studies compared using extraction techniques which are based on MeSH descriptors versus other techniques that are based on extraction from titles, abstracts, or the full-text. Other studies focused on the development of open-source literature mining tool. Some tools were developed mainly to detect the relationships between MeSH descriptors in MEDLINE.
In the following section a sample from different the efforts done in the development of different tools, packages and frameworks will be represented and explained in brief.
LITERATURE MINING TOOLS, PACKAGES, and FRAMEWORKS
2.1 PubMedMineR
PubMedMineR is an R package with text-mining algorithms to analyze PubMed abstracts [2]. It can be used to figure out MeSH-based Associations in PubMed [3]
The PubMed.MineR tool and its documentation are available here or from here.
The tool offers a different set of functionalities that allows the user to read and retrieve information (in XML format). Other sets of functions are entitled to classify documents into categories or provide automatic summarization of a set of documents. Another function is recognizing and normalizing named entities (like genes, proteins or drugs).
The workflow includes 4 phases: searching and retrieving; (2) filtering MeSH descriptors (using UMLs); (3) Statistical reports generation (for UMLs used and MSH descriptors) and, (4) Figuring out association rules between filtered MeSH descriptors.
The output can be displayed in different formats (as plain text, XML, or PDF).
2.2 MeSHmap
This tool aims to exploit the MeSH indexing related to MEDLINE records. MeSHmap has different features: (1) search via PubMed; (2) user-driven exploration of the MeSH terms and subheadings in the result. The visionary plan of the tools includes promising features like comparing entities of the same category (drugs or procedures) and generate maps for the entities where the relationship between two entities in the map is proportional to the degree of similarity in the MeSH metadata of the MEDLINE documents [4].
2.3 MedlineR
MedlineR is an open source library written in R language and used for the data mining of the Medline literature [5].
It entails different functions that could query the NCBI PubMed database, build the co-occurrence matrix, and help in the visualization of the network topology of the query terms.
Users can add their inventions to extend the functionalities of the library using R language. Bioinformaticians are invited to develop more and more tools based-on or using MedlineR.
2.4 Data Mining with Meva in MEDLINE
Medline Evaluator (Meva) is a medical scientific data-mining web service for analyzing the bibliographic fields returned by a PubMed query [6].
In Meva, results are well represented graphically representing counts and relations of the fields using different graphical and statistical methodologies (histograms, correlation tables, detailed sorted lists, or MeSH trees).
Advanced features include applying filters to limit the analysis in the mining process. The output can be populated in different formats (HTML or in a delimited format). The results can then be either be printed, imported to medical databases, or displayed offline.
You can try Meva here.
2.5 PubMedPortable
PubMedPortable is a framework used to support the development of text mining applications [7].
There are many tools that could identify named entities (review section 2.1).
Although PubMed is a huge biomedical literature database, there is no known way to apply Natural Language Processing (NLP) tools or connect them to it. So, there is a great need for a data environment where different applications can be combined to develop text mining applications.
PubMedPortable builds a relational in-house database from a full-text index based-on PubMed articles and citations. It provides an interoperable environment where different NLP approaches can be applied in different programming languages. In addition, queries can be run on several operating systems without facing any problems.
The software can be downloaded here for free (for Linux). For other operating systems you must use a virtual container.
If you want to try any of the previous tools to be trained on querying PubMed, you can use John Snow Labs healthcare data sets that include the “MEDLINE PubMed Journal Citation Database” to validate your results.
This dataset contains NLM’s database of citations and abstracts in the fields of medicine, nursing, dentistry, veterinary medicine, health care systems, and preclinical sciences.
Mining the PubMed database provides researchers with a wealth of information that can enhance clinical practice and patient outcomes through systematic reviews and meta-analyses. By leveraging Generative AI in Healthcare and deploying a Healthcare Chatbot, healthcare professionals can streamline the retrieval of relevant studies, improve patient interaction, and ensure that clinical decisions are informed by the most current and comprehensive evidence available.