Voice of Patients (VoP) NER, a brand-new Named Entity Recognition (NER) model released by John Snow Labs, can extract clinical entities from patient forums much better than any other clinical NER model.

VOP NER Model Sample Extraction
The Need for Voice of the Patients NER Model
Even if there are many other clinical named entity recognition (NER) models out there, extracting clinical entities from health forums can be a challenging task due to the reasons stated below:
- Informal language: As stated in the explanation, the language used in medical forums can be more informal and conversational than in formal medical documents. This makes it challenging for classical NER models to accurately extract named entities from patient text data, as the language may not follow typical grammatical and syntax rules.
- Domain-specific terminology: The terminology used in patient text data may be specific to certain medical conditions or diseases, making it challenging for classical NER models to accurately identify named entities. For instance, a medical term like “idiopathic thrombocytopenic purpura” may not be immediately recognizable by a classical NER model unless it has been specifically trained on this type of terminology.
- Variability: Patients may describe their symptoms and experiences with medical treatments in different ways, making it difficult for classical NER models to recognize and extract named entities consistently.
- Labeled data availability: Medical forums can be a valuable source of patient text data, but labeled data for named entities related to patient health and medical issues may be scarce. A targeted VoP NER model trained on relevant datasets can help to address this issue by providing labeled data for named entities related to patient health and medical issues.
- Clinical relevance: Named entities related to patient health and medical issues can provide valuable insights into patient experiences, symptoms, and treatment outcomes. Accurately identifying and extracting these named entities can help clinicians develop better treatment plans and improve patient outcomes. A targeted VoP NER model can assist in identifying these named entities accurately and efficiently, thus improving the quality of patient care.
Voice of the Patients NER Model
The Spark NLP Voice of The Patients NER (named entity recognition) model is a machine learning model that has been trained to automatically identify and extract named entities from text data related to patient health and medical issues. The model has been specifically trained on text data that has been scraped from medical forums, where patients may discuss their health conditions, symptoms, and experiences with medical treatments.
The main difference between datasets from medical forums and formal medical documents is the language used. Formal medical documents are typically written in technical and professional language that is used by medical experts and practitioners, whereas the language used in medical forums can be more informal and conversational. So, the model that is trained on formal medical data may not be effective in everyday patients’ speaking.
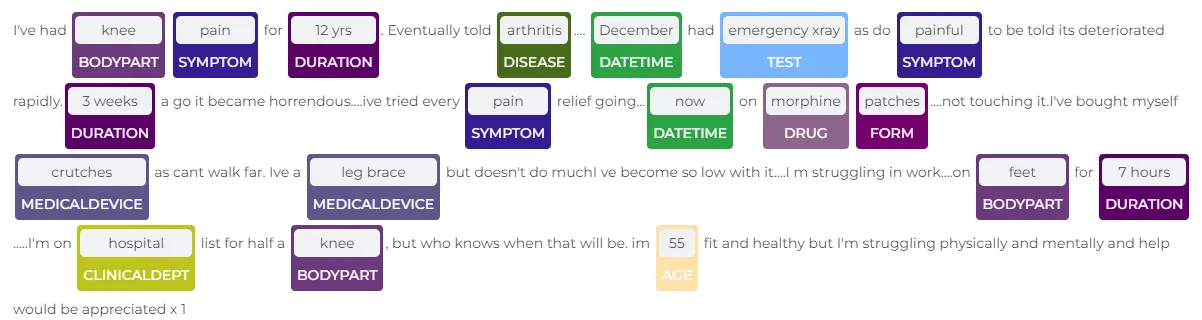
VOP NER Model Sample Extraction
With the VoP NER model, it is possible to automatically identify and extract key information from patient text data, such as the specific body part being discussed, the type of drug being referenced, or the symptoms being experienced. This information can be used in a variety of applications, such as improving patient diagnosis, tracking the spread of diseases, or identifying trends and patterns in patient health data. Here are the current VOP entities:
+--------------------+-------------------------+---------------------+ | BodyPart | Vaccine | AdmissionDischarge | | Drug | Disease | Test | | Dosage | VitalTest | Laterality | | ClinicalDept | Strength | Symptom | | DateTime | InjuryOrPoisoning | Duration | | RelationshipStatus | Frequency | Age | | Employment | Route | Gender | | Form | PsychologicalCondition | Procedure | | Disease | Dosage_Strength | | +--------------------+-------------------------+---------------------+
The Spark NLP VoP model is currently under development and a WIP (work in progress) and slim (with fewer entities) version has just been released. The annotation and training process continues. The final release will include more entities and is expected to have better performance.
Here is a pipeline that can be used to extract NER entities using the VoP NER model:
document_assembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentence_detector = SentenceDetectorDLModel.pretrained("sentence_detector_dl", "en")\
.setInputCols(["document"])\
.setOutputCol("sentence")
tokenizer = Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")
clinical_embeddings = WordEmbeddingsModel.pretrained("embeddings_clinical", "en", "clinical/models")\
.setInputCols(["sentence", "token"])\
.setOutputCol("embeddings")
ner_model = MedicalNerModel.pretrained("ner_vop_slim_wip", "en", "clinical/models")\
.setInputCols(["sentence", "token","embeddings"])\
.setOutputCol("ner")
ner_converter = NerConverterInternal()\
.setInputCols(["sentence", "token", "ner"])\
.setOutputCol("ner_chunk")
pipeline = Pipeline(stages=[
document_assembler,
sentence_detector,
tokenizer,
clinical_embeddings,
ner_model,
ner_converter
])
sample_texts = ["""Hello,I'm 20 year old girl. I'm diagnosed with hyperthyroid
1 month ago. I was feeling weak, light headed,poor digestion, panic attacks,
depression, left chest pain, increased heart rate, rapidly weight loss,
from 4 months. Because of this, I stayed in the hospital and
just discharged from hospital. I had many other blood tests, brain mri,
ultrasound scan, endoscopy because of some dumb doctors bcs
they were not able to diagnose actual problem. Finally I got an appointment
with a homeopathy doctor finally he find that i was suffering from hyperthyroid
and my TSH was 0.15 T3 and T4 is normal . Also i have b12 deficiency and
vitamin D deficiency so I'm taking weekly supplement of vitamin D and
1000 mcg b12 daily. I'm taking homeopathy medicine for 40 days and
took 2nd test after 30 days. My TSH is 0.5 now. I feel a little bit relief
from weakness and depression but I'm facing with 2 new problem from last week
that is breathtaking problem and very rapid heartrate.
I just want to know if i should start allopathy medicine or homeopathy is okay?
Bcs i heard that thyroid take time to start recover.
So please let me know if both of medicines take same time.
Because some of my friends advising me to start allopathy and never take a chance
as i can develop some serious problems.Sorry for my poor english😐Thank you."""]
data = spark.createDataFrame(sample_texts, StringType()).toDF("text")
result = pipeline.fit(data).transform(data)
Result:
+--------------------+-----+----+----------------------+ |chunk |begin|end |ner_label | +--------------------+-----+----+----------------------+ |<span class="hljs-number">20</span> year old |<span class="hljs-number">10</span> |<span class="hljs-number">20</span> |Age | |girl |<span class="hljs-number">22</span> |<span class="hljs-number">25</span> |Gender | |hyperthyroid |<span class="hljs-number">47</span> |<span class="hljs-number">58</span> |Disease | |<span class="hljs-number">1</span> month ago |<span class="hljs-number">60</span> |<span class="hljs-number">70</span> |DateTime | |weak |<span class="hljs-number">87</span> |<span class="hljs-number">90</span> |Symptom | |panic attacks |<span class="hljs-number">122</span> |<span class="hljs-number">134</span> |PsychologicalCondition| |depression |<span class="hljs-number">137</span> |<span class="hljs-number">146</span> |PsychologicalCondition| |left |<span class="hljs-number">149</span> |<span class="hljs-number">152</span> |Laterality | |chest |<span class="hljs-number">154</span> |<span class="hljs-number">158</span> |BodyPart | |pain |<span class="hljs-number">160</span> |<span class="hljs-number">163</span> |Symptom | |heart rate |<span class="hljs-number">176</span> |<span class="hljs-number">185</span> |VitalTest | |weight loss |<span class="hljs-number">196</span> |<span class="hljs-number">206</span> |Symptom | |<span class="hljs-number">4</span> months |<span class="hljs-number">215</span> |<span class="hljs-number">222</span> |Duration | |hospital |<span class="hljs-number">258</span> |<span class="hljs-number">265</span> |ClinicalDept | |discharged |<span class="hljs-number">276</span> |<span class="hljs-number">285</span> |AdmissionDischarge | |hospital |<span class="hljs-number">292</span> |<span class="hljs-number">299</span> |ClinicalDept | |blood tests |<span class="hljs-number">319</span> |<span class="hljs-number">329</span> |Test | |brain |<span class="hljs-number">332</span> |<span class="hljs-number">336</span> |BodyPart | |mri |<span class="hljs-number">338</span> |<span class="hljs-number">340</span> |Test | |ultrasound scan |<span class="hljs-number">343</span> |<span class="hljs-number">357</span> |Test | |endoscopy |<span class="hljs-number">360</span> |<span class="hljs-number">368</span> |Procedure | |doctors |<span class="hljs-number">391</span> |<span class="hljs-number">397</span> |Employment | |homeopathy doctor |<span class="hljs-number">486</span> |<span class="hljs-number">502</span> |Employment | |he |<span class="hljs-number">512</span> |<span class="hljs-number">513</span> |Gender | |hyperthyroid |<span class="hljs-number">546</span> |<span class="hljs-number">557</span> |Disease | |TSH |<span class="hljs-number">566</span> |<span class="hljs-number">568</span> |Test | |T3 |<span class="hljs-number">579</span> |<span class="hljs-number">580</span> |Test | |T4 |<span class="hljs-number">586</span> |<span class="hljs-number">587</span> |Test | |b12 deficiency |<span class="hljs-number">613</span> |<span class="hljs-number">626</span> |Disease | |vitamin D deficiency|<span class="hljs-number">632</span> |<span class="hljs-number">651</span> |Disease | |weekly |<span class="hljs-number">667</span> |<span class="hljs-number">672</span> |Frequency | |supplement |<span class="hljs-number">674</span> |<span class="hljs-number">683</span> |Drug | |vitamin D |<span class="hljs-number">688</span> |<span class="hljs-number">696</span> |Drug | |<span class="hljs-number">1000</span> mcg |<span class="hljs-number">702</span> |<span class="hljs-number">709</span> |Dosage_Strength | |b12 |<span class="hljs-number">711</span> |<span class="hljs-number">713</span> |Drug | |daily |<span class="hljs-number">715</span> |<span class="hljs-number">719</span> |Frequency | |homeopathy medicine |<span class="hljs-number">733</span> |<span class="hljs-number">751</span> |Drug | |<span class="hljs-number">40</span> days |<span class="hljs-number">757</span> |<span class="hljs-number">763</span> |Duration | |after <span class="hljs-number">30</span> days |<span class="hljs-number">783</span> |<span class="hljs-number">795</span> |DateTime | |TSH |<span class="hljs-number">801</span> |<span class="hljs-number">803</span> |Test | |now |<span class="hljs-number">812</span> |<span class="hljs-number">814</span> |DateTime | |weakness |<span class="hljs-number">849</span> |<span class="hljs-number">856</span> |Symptom | |depression |<span class="hljs-number">862</span> |<span class="hljs-number">871</span> |PsychologicalCondition| |last week |<span class="hljs-number">912</span> |<span class="hljs-number">920</span> |DateTime | |rapid heartrate |<span class="hljs-number">960</span> |<span class="hljs-number">974</span> |Symptom | |thyroid |<span class="hljs-number">1074</span> |<span class="hljs-number">1080</span>|BodyPart | +--------------------+-----+----+----------------------+
Conclusion
Overall, this NER model represents a valuable tool for extracting valuable insights from patient text data related to medical issues and has the potential to drive significant improvements in patient care and medical research. By automating the process of identifying and extracting named entities from text, this model can help to save time and effort for medical professionals, while also enabling new insights and discoveries in the field of medical research.
Healthcare NLP models are licensed, so if you want to use these models, you can watch Get a Free License For John Snow Labs NLP Libraries video and request one from https://www.johnsnowlabs.com/install/.
You can follow us on Medium and Linkedin to get further updates or join slack support channel to get instant technical support from the developers of Spark NLP. If you want to learn more about the library and start coding right away, please check our certification training notebooks.































