Tl; DR: This post explains why specialized pretrained PHI pipelines are often the best starting point for data scientists working with clinical text. Instead of building a custom PHI system from scratch, you can deploy John Snow Labs’ one-liner de-identification pipelines that use state-of-the-art NER models and rule-based solutions to detect and extract sensitive identifiers and generate de-identified output — helping teams move faster while maintaining strong accuracy and supporting privacy-compliant workflows.

John Snow Labs’ latest Healthcare NLP release 6.3.0 includes a wide range of updates — new models, pipeline improvements, library additions, and patches across the stack.
In this post, I’m focusing on one particularly practical upgrade for anyone working with real clinical text: specialized pretrained pipelines for PHI detection and de-identification. These pipelines use state-of-the-art NER models to automatically identify and extract sensitive identifiers in clinical documents, helping teams meet privacy and compliance needs. The key advantage is speed and simplicity: instead of building a PHI system from scratch — collecting data, annotating PHI, training, and maintaining custom models — you can deploy a robust de-identification workflow with one-liner implementations.
For data scientists, this means less time spent on infrastructure and edge-case engineering, and more time on downstream work like cohorting, analytics, information extraction, etc.
This release also includes pipelines that incorporate zero-shot NER stages. These “zero-shot” variants are useful when you want strong PHI coverage out of the box — especially for messy, heterogeneous note styles — without investing in task-specific retraining. In practice, they provide a fast way to test PHI detection behavior and iterate quickly before deciding whether deeper customization is needed.
Below are the seven specialized pretrained PHI pipelines released by John Snow Labs. Each pipeline packages an end-to-end PHI workflow — combining pretrained NER with rule-based components — to detect sensitive identifiers (listed in the Description column) and produce de-identified clinical text. While they share the same goal (privacy-safe documents), they can vary in coverage and aggressiveness depending on note style and PHI formats.
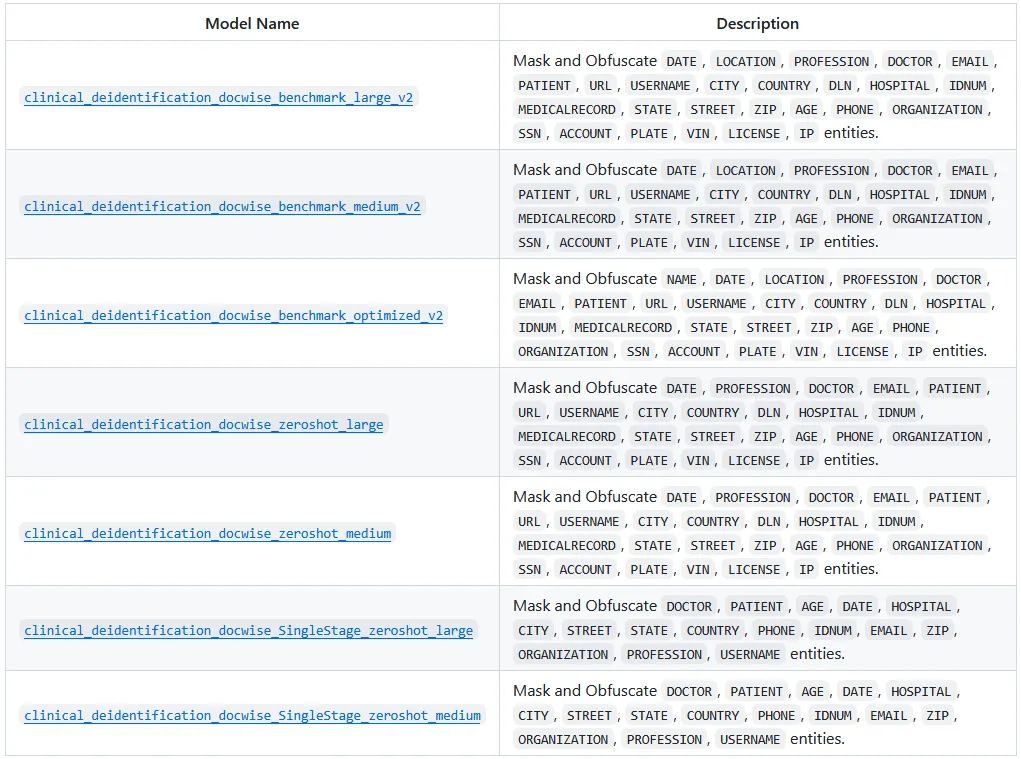
Check the end of the article for the model links.
One-Liner Pretrained Pipelines for PHI Detection and Extraction
At the core, these pipelines combine multiple NER components — both pretrained models and rule-based matchers and parsers — to capture PHI in messy, real-world clinical notes, including identifiers that follow consistent patterns. The pipelines wrap the full flow — document assembly, NER inference, and de-identification — into a deployable unit, making it easy to move from raw notes to privacy-safe text with minimal code and consistent outputs.
In John Snow Labs Healthcare NLP, the pretrained pipelines are packaged so you can load and run them immediately — in a single line:
from sparknlp.pretrained import PretrainedPipeline
deid_pipeline = PretrainedPipeline("clinical_deidentification_docwise_benchmark_large", "en", "clinical/models")
# Try the pipeline on a sample text.
text = """Dr. John Lee, from Royal Medical Clinic in Chicago, attended to the patient on 11/05/2024.
The patient's medical record number is 56467890.
The patient, Emma Wilson, is 50 years old, her Contact number: 444-456-7890 ."""
deid_result = deid_pipeline.fullAnnotate(text)
To illustrate the pipeline behavior, I ran it on a small sample note containing common PHI types (name, location, date, medical record number, phone etc). The pipeline produces two complementary outputs: masking for maximum privacy clarity, and obfuscation to keep the text natural for downstream NLP.

To be more clear, these two outputs serve different needs: masked text is ideal for strict redaction and inspection, while obfuscated text is useful when you want de-identified notes that remain readable for modeling, analytics, or annotation workflows.
Conclusion
PHI handling is one of those steps that can slow down every clinical NLP project — especially when you try to build a solution from scratch. With this release, John Snow Labs introduces specialized pretrained pipelines for PHI detection and de-identification that package strong defaults into a deployable workflow.
By leveraging state-of-the-art NER models (along with pattern-driven components for structured identifiers), these pipelines automatically identify and extract sensitive information and generate de-identified output that supports privacy requirements in real clinical documents.
The practical advantage is simplicity: rather than building and maintaining a custom PHI system, teams can deploy privacy protection with one-liner pipelines that streamline clinical document sanitization while maintaining strong accuracy.
Model Links
- clinical_deidentification_docwise_benchmark_large_v2
- clinical_deidentification_docwise_benchmark_medium_v2
- clinical_deidentification_docwise_benchmark_optimized_v2
- clinical_deidentification_docwise_zeroshot_large
- clinical_deidentification_docwise_zeroshot_medium
- clinical_deidentification_docwise_SingleStage_zeroshot_large
- clinical_deidentification_docwise_SingleStage_zeroshot_medium





























