Developed by John Snow Labs, Healthcare NLP is the leading platform for clinical and biomedical language processing. While new versions roll out every 2–4 weeks, 6.1 stands out as one of the most significant releases to date.
This release introduces brand-new medical multimodal LLMs along with lightweight, text-only LLMs that can run on standard, commodity hardware, making advanced clinical NLP accessible to a broader range of users. Together, these innovations mark a major step forward in both performance and usability for the healthcare AI community.
Healthcare NLP 6.1: What’s New
Medical Vision LLM Module for Multimodal AI
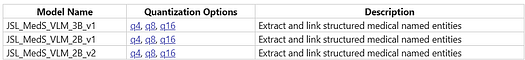
Advancing clinical AI with integrated visual–language understanding, the new MedicalVisionLLM annotator extends LLM capabilities to process both text and medical images. This major addition introduces 9 new Vision-Language Models (VLMs) for diagnostic image interpretation, image-to-text summarization, clinical entity extraction, and integrated clinical documentation analysis. Built on the latest multimodal AI advances, it enables richer clinical insights and informed decision-making across medical workflows.
These models are available in the John Snow Labs Models Hub.
Example for an Image File:
The code snippet below lets you extract medical details from the image file.
prompt = """Extract demographics, clinical disease and medication informations"""
input_df = vision_llm_preprocessor(
spark=spark,
images_path="./images",
prompt=prompt,
output_col_name="prompt"
)
document_assembler = DocumentAssembler()\
.setInputCol("prompt")\
.setOutputCol("document")
image_assembler = ImageAssembler()\
.setInputCol("image")\
.setOutputCol("image_assembler")
medical_vision_llm = MedicalVisionLLM.pretrained("jsl_meds_vlm_3b_q4_v1", "en", "clinical/models")\
.setInputCols(["document", "image_assembler"])\
.setOutputCol("completions")\
.setChatTemplate("vicuna")\
.setBatchSize(4)\
.setNPredict(-1)\
.setTemperature(0)\
.setNGpuLayers(99)\
pipeline = Pipeline(
stages = [
document_assembler,
image_assembler,
medical_vision_llm
])
Result:
{
"Prescription Date": "2023-08-30",
"Diagnosis": "Malaria",
"Medicine Name": "TAB. ABCIXIMAB",
"Dosage": "1 Morning",
"Duration": "4 Days",
"Follow-up Date": "2023-09-04",
"Doctor Name": "Dr. Akshara",
"Hospital Name": "SMS hospital"
}
Please check the Multi Modal LLMs Notebook for details.
The same models can extract medical entities from text with only a slight change in the pipeline.
Example:
document_assembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
medical_llm = MedicalLLM.pretrained("jsl_meds_ner_vlm_2b_q8_v1", "en", "clinical/models")\
.setInputCols("document")\
.setOutputCol("completions")\
.setBatchSize(1)\
.setNPredict(100)\
.setUseChatTemplate(True)\
.setTemperature(0)
pipeline = Pipeline(
stages = [
document_assembler,
medical_llm
])
med_ner_prompt = """
### Template:
{
"drugs": [
{
"name": "",
"reactions": []
}
]
}
### Text:
I feel a bit drowsy & have a little blurred vision , and some gastric problems .
I 've been on Arthrotec 50 for over 10 years on and off , only taking it when I needed it .
Due to my arthritis getting progressively worse , to the point where I am in tears with the agony.
Gp 's started me on 75 twice a day and I have to take it every day for the next month to see how I get on , here goes .
So far its been very good , pains almost gone , but I feel a bit weird , did n't have that when on 50.
"""
data = spark.createDataFrame([[med_ner_prompt]]).toDF("text")
results = pipeline.fit(data).transform(data)
results.select("completions").show(truncate=False)
Results:
{'drugs': [
{
'name': 'Arthrotec 50',
'reactions': [
'drowsy',
'blurred vision',
'gastric problems'
]
},
{
'name': 'Gp 75',
'reactions': [
'pains almost gone',
'weird'
]
}]
}
With a minor pipeline adjustment, the same models can extract medical entities from multiple document types, including PDFs, Word, Excel, PowerPoint, HTML, Text, Email, and Markdown.
Please review the code below using this sample PDF file.
Here, Reader2Doc annotator will read the pdf file and convert its content into plain text — to be processed in the following pipeline stages.
from sparknlp.reader.reader2doc import Reader2Doc
reader2doc = Reader2Doc() \
.setContentType("application/pdf")\
.setContentPath("pdf_files/*pdf")\
.setOutputCol("raw_text")\
.setExplodeDocs(False)\
.setFlattenOutput(True)\
.setOutputFormat("plain-text")
pipeline = Pipeline(stages=[reader2doc])
empty_df = spark.createDataFrame([], "string").toDF("text")
model = pipeline.fit(empty_df)
result_df = model.transform(empty_df)
result_df.show(truncate=False)
Simply write your prompt and invoke the LLM model in the next step to extract the medical entities from the pdf document:
prompt = """
### Template:
{
"clinical":
[
"disease":[],
"symptom":[]
],
"demographic":
[
"name":[],
"date":[],
"id":[]
]
}
### Text:
"""
medical_llm = MedicalLLM.pretrained("jsl_meds_ner_vlm_2b_q16_v1", "en", "clinical/models")\
.setInputCols("document")\
.setOutputCol("completions")\
.setBatchSize(1)\
.setNPredict(10000)\
.setUseChatTemplate(True)\
.setTemperature(0)\
.setNGpuLayers(100)
The results are quite satisfactory:
{
"clinical": {
"disease": [
"Mesothelioma",
"pleural effusion",
"atrial fibrillaIon",
"anemia",
"ascites",
"esophageal reflux",
"deep venous thrombosis"
],
"symptom": [
"nonproductive cough",
"right-sided chest pain",
"fever",
"right-sided pleural effusion",
"cough with right-sided chest pain",
"pericardiIs",
"pericardectomy",
"atrial fibrillaIon",
"RNCA with intracranial thrombolyIc treatment",
"PTA of MCA",
"Mesenteric venous thrombosis",
"pericardial window",
"cholecystectomy",
"LeZ thoracentesis"
]
},
"demographic": {
"name": [
"Hendrickson, Ora MR."
],
"date": [
"2007-08-24",
"2007-08-20",
"2007-08-31"
],
"id": [
"7194334"
]
}
}
Refer to the Notebook for details.
JSL Medical LLM Collection: New v4 & v5 Models in 4B/8B, q4–q16 Formats
This release adds 9 new models across v4/v5, 4B & 8B sizes, and q4/q8/q16 quantization, balancing speed, memory efficiency, and latency. Optimized for accurate clinical text understanding, they power real-time doctor–patient assistance, patient history summarization, and insight extraction from medical literature and EHRs — with quantized models enabling deployment from GPU clusters to resource-limited settings.
Let’s use a model to extract the symptoms and suggest the most appropriate treatment.
Example:
medical_llm = medical.MedicalLLM.pretrained("jsl_meds_4b_q16_v5", "en", "clinical/models")\
.setInputCols("document")\
.setOutputCol("completions")\
.setBatchSize(1)\
.setNPredict(100)\
.setUseChatTemplate(True)\
.setTemperature(0)
pipeline = nlp.Pipeline(stages=[
document_assembler,
medical_llm
])
prompt = """
A 23-year-old pregnant woman at 22 weeks gestation presents with burning upon urination. She states it started 1 day ago and has been worsening despite drinking more water and taking cranberry extract. She otherwise feels well and is followed by a doctor for her pregnancy. Her temperature is 97.7°F (36.5°C), blood pressure is 122/77 mmHg, pulse is 80/min, respirations are 19/min, and oxygen saturation is 98% on room air. Physical exam is notable for an absence of costovertebral angle tenderness and a gravid uterus.
Which of the following is the best treatment for this patient?
A: Ampicillin
B: Ceftriaxone
C: Ciprofloxacin
D: Doxycycline
E: Nitrofurantoin
"""
Results:
The patient presents with symptoms suggestive of a urinary tract infection (UTI) during pregnancy. Given the gestational age (22 weeks), the most appropriate treatment option is E: Nitrofurantoin.
LLM Architecture Upgrade: Faster, Scalable, More Accurate
The LLM architecture in Healthcare NLP now supports the latest LLM families, boosting GPU inference speed by ~10% for both MedicalLLM and LLMLoader.
It also adds compatibility with new architectures, enabling faster, more scalable deployments and seamless integration with state-of-the-art models for advanced healthcare NLP.
Faster Healthcare Modules: Ongoing Optimization & Benchmarking
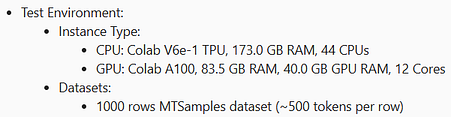
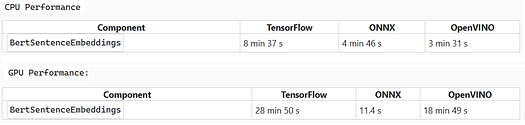
We benchmarked TensorFlow, ONNX, and OpenVINO implementations under identical conditions to compare architectural performance in the healthcare library.
Tests ran on CPU and GPU with the same dataset and batch settings for fairness.
Benchmark settings and environments are shown below:
ONNX outperformed TensorFlow on both CPU and GPU (with massive GPU gains), OpenVINO led CPU speed but trailed ONNX on GPU, and all tests used identical hardware with warm-up runs to prevent cold-start effects.
Future releases will prioritize ONNX and OpenVINO architectures — given their strong GPU (ONNX) and CPU (OpenVINO) performance — to expand model coverage, optimize pipelines, and maximize hardware acceleration compatibility.
7 New Mapper Models for HPO, Genes, Diseases & Biomedical Concepts
These 7 ChunkMapper models provide fast, lightweight lookup layers for key biomedical vocabularies, including genes, diseases, HPO terms, EOM phenotypes, and UMLS concepts.
- gene_disease_mapper: Maps genes to their related diseases.
- gene_hpo_code_mapper: Maps genes to their corresponding HPO codes.
- hpo_code_eom_mapper: Maps HPO codes to their related extraocular movements (EOM).
- hpo_code_gene_mapper: Maps HPO codes to related genes.
- hpo_umls_mapper: Maps HPO codes to corresponding UMLS codes.
- umls_hpo_mapper: Maps UMLS codes to corresponding HPO codes.
- hpo_code_gene_disease_mapper: Maps HPO codes to their associated genes and further maps those genes to related diseases.
Example:
mapperModel = ChunkMapperModel.pretrained("gene_disease_mapper", "en", "clinical/models")\
.setInputCols(["ner_chunk"])\
.setOutputCol("mappings")\
.setRels(["disease"])
model = nlp_pipeline.fit(spark.createDataFrame([['']]).toDF("text"))
result = model.transform(spark.createDataFrame([["We will systematically examine seven genes (CHN1, MDH1, and SNAP25) that are altered in the three neurodegenerative diseases."]]).toDF("text"))
Results:
Domain-Specific Pretrained Clinical Pipelines
This release adds hybrid pretrained pipelines for fast, accurate clinical document analysis.
Each pipeline combines multiple SOTA models into a one-liner solution, removing the need for manual model chaining or workflow setup and enabling rapid, reliable deployment for targeted clinical tasks.
The following pipelines can be used to extract PHI information such as ‘CONTACT’, ‘DATE’, ‘ID’, ‘LOCATION’, ‘PROFESSION’, ‘DOCTOR’, ‘EMAIL’, ‘PATIENT’, ‘URL’, ‘USERNAME’, ‘CITY’, ‘COUNTRY’, ‘DLN’, ‘HOSPITAL’, ‘IDNUM’, ‘LOCATION_OTHER’, ‘MEDICALRECORD’, ‘STATE’, ‘STREET’, ‘ZIP’, ‘AGE’, ‘PHONE’, ‘ORGANIZATION’, ‘SSN’, ‘ACCOUNT’, ‘PLATE’, ‘VIN’, ‘LICENSE’, and ‘IP’ entities:
- ner_docwise_benchmark_medium
- ner_docwise_benchmark_large
- clinical_deidentification_docwise_benchmark_medium
- clinical_deidentification_docwise_benchmark_large
Please refer to the Notebook for details.
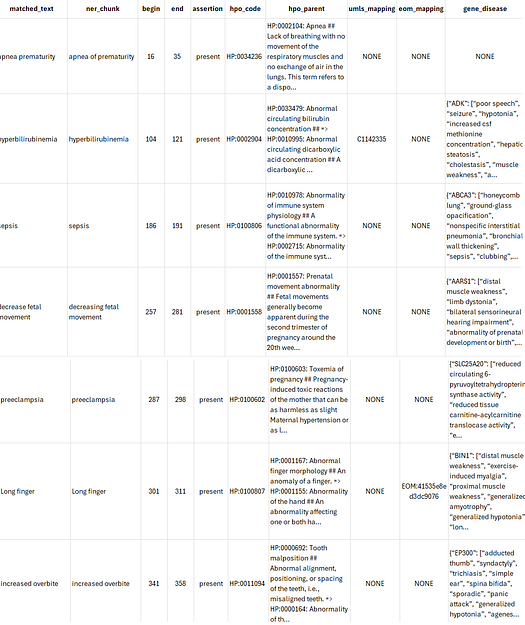
hpo_mapper_pipeline_v3: Designed to extract phenotype-related entities from clinical or biomedical text, map them to their corresponding Human Phenotype Ontology (HPO) codes, and determine their assertion status.
Here’s how the HPO code mapper pipeline performs on a sample text.
from sparknlp.pretrained import PretrainedPipeline
pipeline = PretrainedPipeline("hpo_mapper_pipeline_v3", "en", "clinical/models")
result = pipeline.fullAnnotate("""APNEA: Presumed apnea of prematurity since < 34 wks gestation at birth.
HYPERBILIRUBINEMIA: At risk for hyperbilirubinemia d/t prematurity.
1/25-1/30: Received Amp/Gent while undergoing sepsis evaluation.
Mother is A+, GBS unknown, and infant delivered
for decreasing fetal movement and preeclampsia.
Long finger and toes detected.
he has a increased overbite expression.
""")
Results:
Updated Notebooks for Easier Healthcare NLP Navigation
- New Deidentification Model Evaluation Notebook
- New Metadata Annotation Converter MOOC Notebook
- New Multi Modal LLMs Notebook
- Updated Chunk Mapping Notebook
- Updated Clinical Deidentification Improvement Notebook
- Updated Loading Medical and Open Source LLMs Notebook
We’ve added and updated numerous clinical models and pipelines, strengthening our healthcare offering.
jsl_meds_vlm_2b_q16_v1jsl_meds_vlm_2b_q8_v1jsl_meds_vlm_2b_q4_v1jsl_meds_vlm_2b_q16_v2jsl_meds_vlm_2b_q8_v2jsl_meds_vlm_2b_q4_v2jsl_meds_4b_q16_v4jsl_meds_4b_q8_v4jsl_meds_4b_q4_v4jsl_meds_4b_q16_v5jsl_meds_4b_q8_v5jsl_meds_4b_q4_v5jsl_meds_8b_q16_v4jsl_meds_8b_q8_v4jsl_meds_8b_q4_v4jsl_meds_vlm_3b_q16_v1jsl_meds_vlm_3b_q8_v1jsl_meds_vlm_3b_q4_v1jsl_meds_vlm_2b_q16_v1jsl_meds_vlm_2b_q8_v1jsl_meds_vlm_2b_q4_v1jsl_meds_vlm_2b_q16_v2jsl_meds_vlm_2b_q8_v2jsl_meds_vlm_2b_q4_v2gene_disease_mappergene_hpo_code_mapperhpo_code_eom_mapperhpo_code_gene_mapperhpo_umls_mapperumls_hpo_mapperhpo_code_gene_disease_mapperhpo_mapper_pipeline_v3ner_docwise_benchmark_mediumner_docwise_benchmark_largeclinical_deidentification_docwise_benchmark_mediumclinical_deidentification_docwise_benchmark_large
For all Healthcare NLP models, please check: Models Hub Page.
Conclusion
This release delivers major advancements across Healthcare NLP, including new medical multimodal LLMs, expanded model collections, performance optimizations, pretrained clinical pipelines, and core reliability improvements.
Together, these updates offer faster, more accurate, and more accessible clinical NLP solutions for real-world healthcare applications.
If you would like to check the whole release notes, you can see it here.