RxNorm provides normalized names for clinical drugs and links its names to many of the drug vocabularies commonly used in pharmacy management and drug interaction software.
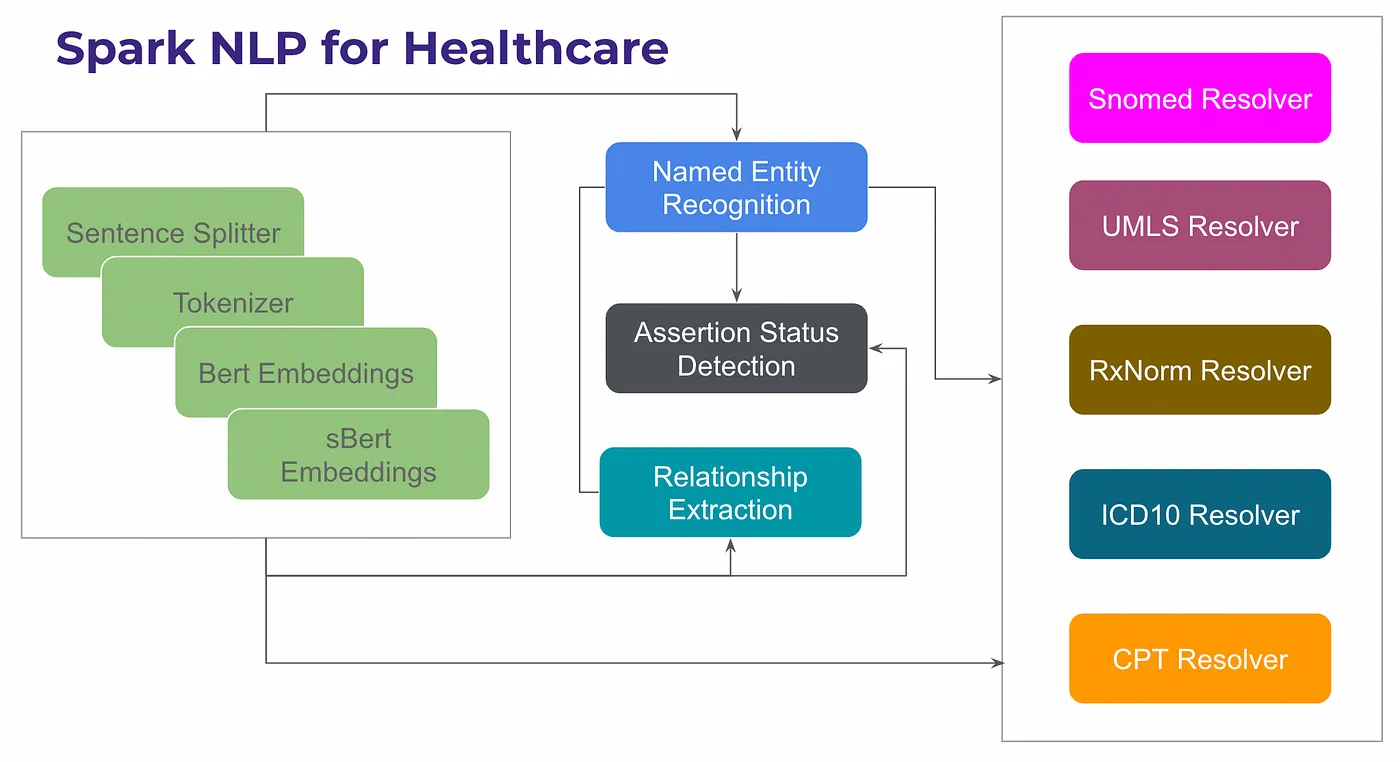
Entity Resolver pipeline in Healthcare NLP
Healthcare NLP v4.3.1 is out! Now you can map clinical entities to RxNorm codes according to the NIH database and map NDC codes to drug brand names.
New Entity Resolver Model for Mapping Rxnorm Codes According To the National Institute of Health (NIH) Database
RxNorm provides normalised names for clinical drugs and links its names to many of the drug vocabularies commonly used in pharmacy management and drug interaction software.
Healthcare NLP comes with 60+ different entity resolver models to support several clinical terminologies (RxNorm, ICD-10-CM, SNOMED, CPT, ATC, HPO, etc.). You can also check entity resolution benchmarks that we got against cloud providers in Comparison of Key Medical NLP Benchmarks — Spark NLP vs AWS, Google Cloud and Azure medium article.
If you want to use sentence entity resolver models, first you need to extract the related clinical entities from clinical texts by using clinical NER models in Spark NLP. You can check Clinical Named Entity Recognition Notebook in Spark NLP Workshop Repo to see how these models can be used for clinical entity extraction. After getting the appropriate entities, you need to get the embeddings of them and feed to the related entity resolver model. There is a Clinical Entity Resolvers Notebook in the same repo that you can find how to implement entity resolver models for several clinical terminologies.
And now, a new sbiobertresolve_rxnorm_nih model is released in v4.3.1 that maps clinical entities and concepts (like drugs/ingredients) to RxNorm codes according to the National Institute of Health (NIH) database using sbiobert_base_cased_mli Sentence Bert Embeddings. You can find an example of this model below.
Example:
...
rxnorm_resolver = SentenceEntityResolverModel.pretrained("sbiobertresolve_rxnorm_nih","en", "clinical/models") \
.setInputCols(["sbert_embeddings"]) \
.setOutputCol("resolution")\
.setDistanceFunction("EUCLIDEAN")
text= "She is given folic acid 1 mg daily , levothyroxine 0.1 mg and aspirin 81 mg daily ."
Result:
| ner_chunk | entity |rxnorm_code | all_codes | resolutions | |:---------------------|:-------|-----------:|:----------------------------------------|:---------------------------------------------------------------------------------| | folic acid 1 mg | DRUG | 12281181 | ['12281181', '12283696', '12270292', ...| ['folic acid 1 MG [folic acid 1 MG]', 'folic acid 1.1 MG [folic acid 1.1 MG]',...| | levothyroxine 0.1 mg | DRUG | 12275630 | ['12275630', '12275646', '12301585', ...| ['levothyroxine sodium 0.1 MG [levothyroxine sodium 0.1 MG]', 'levothyroxine ...| | aspirin 81 mg | DRUG | 12278696 | ['12278696', '12299811', '12298729', ...| ['aspirin 81 MG [aspirin 81 MG]', 'aspirin 81 MG [YSP Aspirin] [aspirin 81 MG ...|
New Chunk Mapper Models for Mapping NDC Codes to Drug Brand Names As Well As Clinical Entities (like drugs/ingredients) to Rxnorm Codes
In Spark NLP, there are 30+ chunk mapper models that were trained for several solutions like mapping clinical terminology codes each other, abbreviations, drug action-treatments etc. and you can find detialed examples in Chunk Mapping Notebook.
Two new chunk mapper models are released in v4.3.1 in addition to 30+ chunk mapper models in Spark NLP.
- ndc_drug_brandname_mapper model maps NDC codes with their corresponding drug brand names as well as RxNorm codes according to the National Institute of Health (NIH).
Example:
...
mapper = ChunkMapperModel.pretrained("ndc_drug_brandname_mapper", "en", "clinical/models")\
.setInputCols("document")\
.setOutputCol("mappings")\
.setRels(["drug_brand_name"])
text= ["0009-4992", "57894-150"]
Result:
| ndc_code | drug_brand_name | |:-----------|:------------------| | 0009-4992 | ZYVOX | | 57894-150 | ZYTIGA |
- rxnorm_nih_mapper model maps entities with their corresponding RxNorm codes according to the National Institute of Health (NIH) database. It returns Rxnorm codes along with their NIH Rxnorm Term Types within a parenthesis.
Example:
...
chunkerMapper = ChunkMapperModel\
.pretrained("rxnorm_nih_mapper", "en", "clinical/models")\
.setInputCols(["ner_chunk"])\
.setOutputCol("mappings")\
.setRels(["rxnorm_code"])
Result:
+-------------------------+-------------+-----------+ |ner_chunk |mappings |relation | +-------------------------+-------------+-----------+ |Adapin 10 MG Oral Capsule|1911002 (SY) |rxnorm_code| |acetohexamide |12250421 (IN)|rxnorm_code| |Parlodel |829 (BN) |rxnorm_code| +-------------------------+-------------+-----------+
All in One Pipeline
Let’s build up a pipeline to show how NER models can be used with sbiobertresolve_rxnorm_nih resolver and rxnorm_nih_mapper chunk mapper models together.
document_assembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentenceDetectorDL = SentenceDetectorDLModel.pretrained("sentence_detector_dl_healthcare", "en", 'clinical/models') \
.setInputCols("document") \
.setOutputCol("sentence")
tokenizer = Tokenizer()\
.setInputCols("sentence")\
.setOutputCol("token")
word_embeddings = WordEmbeddingsModel.pretrained("embeddings_clinical", "en", "clinical/models")\
.setInputCols(["sentence", "token"])\
.setOutputCol("word_embeddings")
clinical_ner = MedicalNerModel.pretrained("ner_posology_greedy", "en", "clinical/models") \
.setInputCols(["sentence", "token", "word_embeddings"]) \
.setOutputCol("ner")
ner_converter_icd = NerConverterInternal() \
.setInputCols(["sentence", "token", "ner"]) \
.setOutputCol("ner_chunk")\
.setWhiteList(['DRUG'])\
.setPreservePosition(False)
c2doc = Chunk2Doc()\
.setInputCols("ner_chunk")\
.setOutputCol("doc_ner_chunk")
sbert_embedder = BertSentenceEmbeddings.pretrained('sbiobert_base_cased_mli', 'en','clinical/models')\
.setInputCols("doc_ner_chunk")\
.setOutputCol("sentence_embeddings")\
.setCaseSensitive(False)
rxnorm_resolver = SentenceEntityResolverModel.pretrained("sbiobertresolve_rxnorm_nih","en", "clinical/models") \
.setInputCols(["sentence_embeddings"]) \
.setOutputCol("rxnorm_code")\
.setDistanceFunction("EUCLIDEAN")
chunkMapper = ChunkMapperModel.pretrained("rxnorm_nih_mapper", "en", "clinical/models")\
.setInputCols(["ner_chunk"])\
.setOutputCol("mappings")\
.setRels(["rxnorm_code"])
resolver_pipeline = Pipeline(
stages = [
document_assembler,
sentenceDetectorDL,
tokenizer,
word_embeddings,
clinical_ner,
ner_converter_icd,
c2doc,
sbert_embedder,
rxnorm_resolver,
chunkMapper
])
Conclusion
Entity mapping is a critical task in clinical NLP and Spark NLP for Healthcare is one of the most popular libraries for this. John Snow Labs is keeping up-to-date this library with new releases every two weeks. There will be new features in the upcoming releases, so keep following us!
Spark NLP for Healthcare models are licensed, so if you want to use these models, you can watch Get a Free License For John Snow Labs NLP Libraries video and request one from https://www.johnsnowlabs.com/install/.
You can follow us on Medium and Linkedin to get further updates or join slack support channel to get instant technical support from the developers of Spark NLP. If you want to learn more about the library and start coding right away, please check our certification training notebooks.































