Oncology has always depended on multidisciplinary tumor boards (MTBs) to interpret complex, multimodal data, such as pathology reports, radiology findings, genomic profiles, prior treatments, patient history, and synthesize this information into personalized cancer treatment recommendations. But the volume and complexity of data now overwhelm traditional review processes.
Roche confronted this challenge directly: matching oncology patients with appropriate National Comprehensive Cancer Network (NCCN) clinical guidelines from comprehensive datasets spanning genetic, epigenetic, and phenotypic information. Their healthcare-specific LLM implementation demonstrates that automated guideline matching can enhance precision in oncology care, ensuring each patient receives tailored treatment recommendations based on latest evidence without overwhelming clinical teams with literature review.
The technical requirements are unforgiving. Comprehensive genomic profiling generates hundreds of variants requiring interpretation. Advanced imaging produces terabytes of data. Clinical notes document treatment sequences across multiple providers. Human tumor boards cannot process this data volume within consultation timeframes while maintaining up-to-date knowledge of evolving guidelines, emerging biomarkers, and active clinical trials.
This article examines what organizations learn when implementing medical LLMs to support tumor board workflows, the evidence from systematic evaluations, and how validated approaches combine specialized language models with human oversight to deliver more consistent, evidence-based oncology care.
Why tumor boards need decision support—and why general-purpose AI falls short
Health systems implementing tumor board AI support consistently identify three recurring challenges that define technical requirements:
Challenge 1: Data integration across heterogeneous sources
MiBA’s experience processing oncology data from electronic health records revealed the operational complexity. Their AI-enhanced pipeline handles 1.4 million physician notes and approximately 1 million PDF reports, including pathology reports, radiology findings, genetic test results, and treatment histories, all with different formats, terminologies, and structures.
The system achieved 93% F1-score for entity extraction and 88% F1-score for relationship extraction, validating that specialized NLP can accurately identify temporal sequences (diagnosis dates, treatment timelines, disease progression), anatomical details (tumor location, metastatic sites), and treatment-related entities (drug regimens, surgical procedures, radiation doses) across unstructured clinical documentation.
The lesson: tumor board support requires extracting structured information from unstructured sources at high accuracy. Generic language models trained on general text miss clinical terminology, abbreviations, treatment protocols, and context-dependent oncology concepts that specialized models recognize.
Challenge 2: Clinical guideline matching at decision-relevant precision
Roche’s implementation demonstrates guideline alignment in practice. Their healthcare-specific LLM analyzes comprehensive patient data to match individual profiles with NCCN clinical guidelines, the evidence-based standards governing oncology treatment decisions. The system “enhances precision in oncology care by ensuring each patient receives tailored treatment recommendations based on the latest guidelines.”
But accuracy requirements are unforgiving. Incorrect biomarker interpretation, missed contraindications, or outdated guideline versions can lead tumor boards to inappropriate recommendations. Systematic physician evaluation showed medical doctors preferring specialized medical LLMs 47% to 25% over GPT-4o on clinical summarization tasks, the core capability tumor boards require for synthesizing patient data. For clinical information extraction (diagnoses, treatments, biomarkers from notes), physicians preferred specialized healthcare NLP 35% to 24%, demonstrating that domain-specific training delivers the accuracy clinical workflows demand.
The lesson: general-purpose LLMs lack the precision oncology decisions require. The 22-point physician preference gap for clinical summarization and 11-point gap for information extraction separate systems suitable for decision support from those requiring extensive manual correction.
Challenge 3: Clinical trial matching from incomplete structured data
MiBA’s pipeline demonstrates why clinical trial matching depends on unstructured data extraction. Their system achieved particularly high accuracy “when cancer staging and biomarker findings are important inclusion criteria as these are often missing from structured EMR data but can be recovered using NLP” from physician notes, pathology reports, and radiology findings.
Cancer staging often appears only in free-text summaries. Biomarker results reside in pathology PDFs. Prior treatment sequences span multiple provider notes. Structured EHR fields miss the context tumor boards need to evaluate trial eligibility. MiBA’s approach of combining NLP entity extraction with LLM reasoning enabled “accurate matching of patients to clinical trial enrollment criteria with high accuracy.”
The lesson: effective tumor board support requires extracting clinical context from unstructured documentation, not just querying structured database fields. Organizations building cancer data registries with LLMs report similar findings, disease response classification and sites of metastases often require NLP extraction from clinical notes for accurate registry population.
What systematic evaluation reveals about medical LLMs in tumor board workflows
Recent research provides systematic evidence for medical LLM capabilities in oncology decision support, revealing both validated strengths and persistent limitations:
Finding 1: Specialized models match human tumor board recommendations in standard cases
A retrospective study of multidisciplinary tumor board decisions found GPT-4 achieving approximately 90% compatibility with human board recommendations across standard oncology scenarios. Another real-world evaluation using ChatGPT 4.0 for 20 anonymized cases found the LLM producing more therapeutic proposals per case than human MTBs (median 3 versus 1) while reaching comparable information depth.
However the CLEVER study’s blind, randomized assessment by practicing medical doctors showed specialized medical LLMs achieving 47% physician preference versus GPT-4o’s 25% on clinical summarization across 500 novel test cases. For biomedical research question-answering, the literature synthesis capability tumor boards require, the specialized medical model “outperforms GPT-4o by a substantial margin” in factuality.
However, even with 90% overall agreement, individual recommendations diverge on rare cancers or edge cases. A sarcoma tumor board evaluation using ChatGPT-4o found “moderate effectiveness” with surgical recommendations, highlighting that complexity and rarity affect performance consistency.
Finding 2: Retrieval-augmented approaches enable guideline currency
Roche’s NCCN guideline matching demonstrates a critical architectural pattern: combining LLM reasoning with retrieval-augmented generation (RAG) over current clinical evidence. Rather than relying solely on pre-trained knowledge, RAG-enabled systems fetch up-to-date guidelines, clinical trial protocols, and literature as part of recommendation generation.
The MEREDITH system, designed to provide tailored treatment recommendations, similarly combines LLM reasoning with retrieval over clinical evidence and molecular data. This architectural choice addresses a fundamental limitation: pre-trained models lack knowledge of guidelines updated after training, newly approved therapies, or recently opened clinical trials. Retrieval-augmented approaches that dynamically incorporate current evidence overcome this constraint.
Finding 3: Efficiency gains are substantial but require validation workflows
The ChatGPT 4.0 tumor board study reported median preparation time of approximately 15 minutes per case versus approximately 35 minutes for human-only review, nearly 2× speedup. This frees oncologists to focus on deliberation, patient communication, and complex cases while LLMs handle literature review, data aggregation, and initial recommendation drafting.
However, efficiency gains require human validation workflows. MiBA’s approach of combining automated extraction with expert review demonstrates the pattern: NLP/LLM systems process large document volumes at speed, but oncologists validate outputs before clinical use. Organizations implementing regulatory-grade oncology data curation emphasize that “the key is the uncompromising quality of regulatory grade data”, efficiency without accuracy compromises patient care.
Technical requirements for medical LLM integration in tumor boards
Based on implementations processing millions of oncology documents and systematic physician evaluations, several technical requirements emerge as non-negotiable:
Requirement 1: Domain-specific training on clinical oncology data
The physician preference gaps, 47% versus 25% for clinical summarization, 35% versus 24% for information extraction, demonstrate that specialized medical training fundamentally changes model performance. Generic models trained on general text fail at clinical entity recognition, oncology terminology, treatment protocol interpretation, and biomarker annotation.
MiBA’s 93% entity extraction and 88% relationship extraction F1-scores validate that models trained on oncology-specific data achieve the precision clinical workflows require. Their pipeline processes pathology reports (tumor type, grade, stage, margins), radiology findings (lesion characteristics, disease extent), genetic reports (mutations, expression levels, pathway alterations), and physician notes (treatment rationale, patient response, complications), each requiring domain-specific language understanding.
Requirement 2: Multi-modal data integration across clinical sources
Tumor board decisions require synthesizing information from multiple modalities: text reports, structured lab values, imaging studies, genomic data. Multimodal oncology AI agents represent the architectural direction, but operational implementations currently rely on extracting information from each modality into structured formats that LLMs can reason over.
Roche’s patient timeline construction demonstrates this integration. Their system extracts and synthesizes chemotherapy treatment data from “diverse clinical notes, including those from primary care providers, oncologists, discharge summaries, emergency departments, pathology, and radiology reports”, creating longitudinal patient views that tumor boards use to evaluate treatment sequences, disease progression, and trial eligibility.
Requirement 3: Retrieval-augmented generation for guideline currency
Pre-trained knowledge becomes stale as guidelines evolve, trials open and close, and new therapies gain approval. MEREDITH’s architecture combining LLM reasoning with retrieval over clinical evidence and molecular data demonstrates the solution: dynamically fetch current guidelines, trial protocols, and literature during recommendation generation.
This approach also addresses evidence transparency requirements. Rather than generating recommendations from opaque model weights, RAG systems cite specific guidelines, trial protocols, or publications, enabling tumor board members to verify reasoning and assess evidence quality. Some implementations embed evidence levels (1A, 2B, etc.) with recommendations, helping oncologists calibrate confidence.
Requirement 4: Human-in-the-loop validation and override
Every implementation emphasizes the same principle: LLM outputs serve as decision support, not autonomous decisions. MiBA’s framework, COTA’s regulatory-grade approach, and academic evaluations all maintain expert review, validation, and final decision authority with oncologists.
The head and neck cancer MTB evaluation found performance varying by specialty, surgical recommendations scored highest while other treatment modalities showed more variability. This heterogeneity reinforces that human experts must review LLM suggestions, calibrate against patient context (comorbidities, preferences, prior treatments, performance status), and make final management decisions.
Requirement 5: Audit trails and provenance tracking
For medico-legal safety and clinical trust, tumor board AI systems must provide transparent reasoning with source citations. The ChatGPT 4.0 study noted that LLMs occasionally suggest therapies based on weaker or preclinical evidence, highlighting the need for systems that explicitly indicate evidence strength and guideline adherence.
Audit requirements include: which data sources informed recommendations, which guidelines were consulted, what evidence level supports each suggestion, when the recommendation was generated (model version, knowledge cutoff), and how human reviewers modified or overrode AI suggestions. Organizations pursuing regulatory-grade quality maintain complete provenance from source documents through extraction, reasoning, recommendation, and final tumor board decision.
Implementation pathway: from pilot to operational tumor board support
Organizations successfully implementing medical LLM support for tumor boards follow a similar maturity progression:
Phase 1: Pilot with retrospective case validation
Select representative tumor types (common cancers with established guidelines) and process historical cases where tumor board decisions are documented. Compare LLM recommendations against actual board decisions. MiBA’s approach of achieving 93% entity extraction provides the accuracy benchmark, systems must reliably extract diagnoses, staging, biomarkers, prior treatments before generating recommendations.
Roche’s NCCN guideline matching demonstrates the validation approach: ensure LLM recommendations align with guideline-concordant options the tumor board selected. Measure not just overall agreement but also clinically significant discrepancies (contraindicated suggestions, missed guideline updates, incorrect trial eligibility).
Key activities: accuracy validation on historical cases, infrastructure setup, integration with EHR/oncology systems, tumor board feedback on recommendation format and evidence presentation.
Phase 2: Prospective pilot with human review (3-4 months)
Introduce LLM-generated case summaries and recommendations into actual tumor board workflows, but as supplemental information reviewed alongside traditional preparation. Oncologists validate accuracy, assess clinical utility, and provide feedback on recommendation quality.
The 15-minute versus 35-minute preparation time reduction observed in published studies represents the efficiency target. However, organizations must validate that speed gains do not compromise decision quality, hence the prospective pilot with full human oversight before operational deployment.
Key activities: real-time case processing, tumor board member training, feedback collection, refinement of recommendation formatting, validation of literature citations and guideline references.
Phase 3: Operational deployment with continuous monitoring (ongoing)
Expand to full tumor board case volume with LLM support as standard workflow component. Maintain human review and override authority while tracking recommendation adoption rates, discrepancy patterns, and clinical outcomes. Implement feedback loops where tumor board corrections refine the system.
A 2025 systematic review identified 56 studies covering 15 cancer types where LLMs supported oncology decision workflows, demonstrating that the approach scales across tumor types when properly validated. However, most studies remain retrospective or limited cohorts; prospective trials with safety monitoring and longitudinal outcomes are still emerging.
Key activities: full-scale deployment, continuous accuracy monitoring, outcome tracking, guideline update incorporation, model retraining on institutional cases, medico-legal framework maintenance.
What operational tumor board LLM support looks like in practice
Organizations implementing medical LLM support consistently converge on similar workflow patterns:
Pre-meeting preparation: AI systems aggregate patient history from multiple EHR sources, extract relevant information from radiology and pathology reports, identify genomic alterations and biomarkers, summarize prior treatment sequences, retrieve comorbidities and performance status, presenting a concise, standardized case summary to the tumor board. Roche’s approach of building “detailed oncology patient timelines” from diverse clinical notes demonstrates this data integration.
Guideline-concordant recommendation generation: The system proposes ranked therapeutic and diagnostic options aligned with NCCN guidelines or institutional protocols. Roche’s LLM “accurately aligns individual patient profiles with the most relevant clinical guidelines,” ensuring recommendations reflect current evidence standards. Systems also identify applicable clinical trials based on eligibility criteria matching, MiBA’s approach of extracting cancer staging and biomarkers enables accurate trial matching where structured data alone would miss eligible patients.
Evidence presentation with transparency: Rather than opaque suggestions, systems present recommendations with rationale, evidence levels, guideline citations, and literature references. The MEREDITH system’s approach of combining LLM reasoning with retrieval-augmented generation over clinical evidence demonstrates this transparency, tumor boards see not just what the system recommends but why and based on which sources.
Human deliberation and final decision: Oncologists, pathologists, radiologists, and other specialists review AI-generated summaries and recommendations, calibrate against patient context (comorbidities, prior toxicities, patient preferences, social determinants, resource availability), and make final management decisions. The 90% compatibility rate between LLM and human recommendations in standard cases does not eliminate deliberation; it makes deliberation more efficient by handling data synthesis and literature review.
Documentation and follow-up: Systems auto-generate tumor board summaries, track recommended follow-up tasks (imaging schedules, lab monitoring, therapy administration), flag care gaps, and ensure scheduling, reducing administrative burden and missed follow-ups that compromise outcomes.
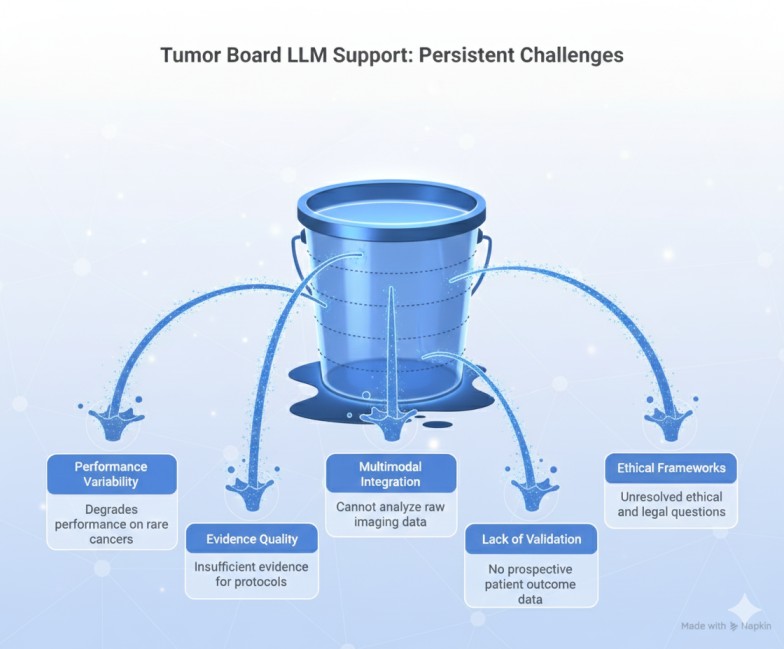
Current limitations and what remains unresolved
Despite promising results, organizations implementing tumor board LLM support encounter persistent challenges:
Limitation 1: Performance variability on rare cancers and edge cases
The sarcoma tumor board evaluation finding “moderate effectiveness” for complex sarcoma cases highlights that rarity and complexity degrade performance. Even the 90% compatibility study noted individual recommendation divergence on rare cancers. Training data for uncommon tumor types is scarce; guidelines for rare cancers often lack high-level evidence; and clinical experience with edge cases cannot be easily encoded.
Organizations address this through human oversight and selective deployment: use LLM support for common cancers with robust guidelines (lung, breast, colon) while maintaining traditional review for rare tumors requiring specialized expertise.
Limitation 2: Evidence quality and guideline adherence variability
The ChatGPT 4.0 study observed that LLMs occasionally suggest therapies based on weaker or preclinical evidence that may not reflect best practice. While LLMs often propose clinical trial options or off-label therapies” potentially expanding choices for patients”, they may also suggest treatments with insufficient evidence for institutional protocols.
Retrieval-augmented approaches that explicitly indicate evidence levels (NCCN Category 1, 2A, 2B, 3) partially address this, but ensuring recommendations match institutional risk tolerance and resource availability requires human calibration.
Limitation 3: Multimodal integration remains incomplete
Current implementations largely process textual summaries rather than raw imaging (CT, MRI, PET scans) or whole slide pathology images. Multimodal oncology agents that directly analyze imaging and pathology are emerging, with specialized tools addressing specific components of this challenge.
Visual NLP demonstrates one validated approach: processing scanned documents, PDFs, DICOM imaging metadata, and handwritten clinical notes that tumor boards frequently encounter. Pathology reports often arrive as scanned PDFs rather than structured text. Radiology findings may include annotated images with embedded measurements. External provider records come in varied formats requiring optical character recognition and layout analysis before information extraction.
Novartis’s implementation of Visual NLP for clinical trial documentation demonstrates the capability at scale: their system processes documents through OCR-based text extraction, language detection, layout and content-based classification, and entity extraction, handling over 40 different document types including handwritten text and date extraction. While this addresses document processing heterogeneity, full multimodal integration, directly analyzing radiologic images for tumor characteristics or pathology slides for cellular features remains an emerging capability not yet operationally deployed in most tumor board workflows.
Until complete multimodal capability matures, LLMs depend on radiologists’ and pathologists’ reports rather than interpreting primary imaging and histologic data, limiting their ability to identify discrepancies between reported findings and raw images, or to detect subtle features that human reviewers might note but not explicitly document in reports.
Limitation 4: Lack of prospective validation with patient outcomes
Most studies remain retrospective comparisons of LLM recommendations against historical tumor board decisions. The systematic review of 56 studies covering 15 cancer types represents substantial evidence, but prospective trials measuring patient outcomes (response rates, progression-free survival, overall survival, quality of life) for LLM-supported versus traditional tumor boards are not yet available. Until such studies demonstrate non-inferiority or superiority on clinical endpoints, adoption remains cautious.
Limitation 5: Ethical, legal, and governance frameworks
Use of AI in treatment decisions raises unresolved questions: Who bears liability when AI-suggested therapy causes harm? How is patient consent obtained for AI-augmented decisions? How are algorithmic biases identified and mitigated? What data governance protects patient privacy when clinical notes train LLMs? Institutional review boards, legal counsel, and clinical leadership must address these questions before widespread operational deployment.
Strategic value: why tumor board LLM support matters beyond efficiency
Organizations investing in medical LLM support for tumor boards realize benefits extending beyond preparation time reduction:
Democratizing access to guideline-concordant care
Not all hospitals maintain large, specialized tumor boards with expertise across tumor types and molecular subtypes. Roche’s NCCN guideline matching approach enables smaller centers to access evidence-based recommendations aligned with national standards, reducing practice variation and ensuring patients receive guideline-concordant options regardless of institutional resources. AI-driven support helps democratize precision oncology for resource-constrained centers.
Expanding therapeutic horizons through trial matching
MiBA’s clinical trial matching from unstructured data addresses a persistent challenge: identifying trial-eligible patients from incomplete structured records. Their approach of extracting cancer staging and biomarkers from clinical notes “enhances performance when these are important inclusion criteria”, connecting patients to experimental therapies they might otherwise miss. LLM systems that automatically identify applicable trials “potentially expand choices for patients” beyond standard-of-care options.
Maintaining knowledge currency as evidence evolves
Oncology guidelines update quarterly. New biomarkers gain clinical utility. Therapies receive expanded indications. FDA approvals alter treatment algorithms. Human tumor boards struggle to maintain currency across specialties. Retrieval-augmented LLM systems that dynamically fetch current guidelines, recent literature, and active trials help boards incorporate emerging evidence without requiring individual members to review every update.
Creating institutional knowledge infrastructure
Organizations implementing tumor board LLM support build reusable knowledge infrastructure. Cancer registry curation systems, longitudinal patient timeline construction, biomarker annotation pipelines, and trial eligibility matching serve not just tumor boards but also quality reporting, outcomes research, and clinical trial recruitment. The investment in structured oncology data extraction compounds benefits across multiple workflows.
Conclusion
Medical LLMs are no longer experimental tools in oncology. They are becoming operational components of tumor board workflows. Roche’s NCCN guideline matching achieving tailored recommendations, MiBA’s pipeline processing 1.4 million notes with 93% entity extraction accuracy, and systematic evaluations showing 90% compatibility with human tumor board decisions demonstrate that specialized medical language models can support, and in some dimensions enhance multidisciplinary cancer care.
But this is not about replacing oncologists or tumor boards. It is about augmenting them with systems that handle data synthesis, literature review, and guideline alignment, freeing human experts to focus on clinical judgment, patient context, and shared decision-making. The 47% versus 25% physician preference for specialized medical LLMs on clinical summarization, and 35% versus 24% for information extraction, validate that domain-specific training delivers accuracy clinical workflows require.
Oncology’s AI moment has arrived, but with constraints. Performance remains variable on rare cancers. Evidence quality requires human calibration. Multimodal integration is incomplete. Prospective outcome trials are pending. Ethical and governance frameworks lag technology capability.
Organizations implementing tumor board LLM support follow a deliberate pathway: retrospective validation on historical cases, prospective pilots with human oversight, operational deployment with continuous monitoring, and feedback loops where tumor board corrections refine the system. The technology is production-ready for common cancers with robust guidelines. The question is whether institutions can implement it responsibly, with proper validation, transparent reasoning, human oversight, and outcomes measurement.
For oncology leaders, informatics teams, and tumor board directors, the path forward requires balancing innovation with caution: leverage medical LLM capabilities where evidence supports them (data integration, guideline matching, trial identification), maintain human authority for final decisions, track outcomes to validate benefit, and build governance frameworks that ensure AI augments rather than compromises cancer care quality.
FAQs
How do specialized medical LLMs compare to general-purpose models like GPT-4 for tumor board support?
Systematic physician evaluation showed medical doctors preferring specialized medical LLMs 47% to 25% over GPT-4o on clinical summarization tasks and 35% to 24% on clinical information extraction, the core capabilities tumor boards require. MiBA’s oncology pipeline achieved 93% F1-score for entity extraction and 88% for relationship extraction processing 1.4 million physician notes, demonstrating that domain-specific training delivers accuracy clinical workflows demand. The CLEVER study’s blind evaluation by practicing medical doctors across 500 novel clinical test cases validated that specialized models “outperform GPT-4o by a substantial margin” on biomedical research question-answering, the literature synthesis capability tumor boards require.
What accuracy has been demonstrated for clinical trial matching using medical LLMs?
MiBA’s AI-enhanced oncology pipeline demonstrates “accurate matching of patients to clinical trial enrollment criteria with high accuracy,” particularly when cancer staging and biomarker findings serve as inclusion criteria, data elements often missing from structured EMR fields but recoverable through NLP extraction from clinical notes. Their system processes physician notes, radiology reports, pathology findings, and genetic reports to extract temporal, anatomical, oncological, pathological, and treatment-related entities that determine trial eligibility. Organizations building cancer data registries report similar capabilities for disease response classification and sites of metastases identification.
Can LLM-generated recommendations replace the need for multidisciplinary tumor boards?
No. Retrospective evaluation found LLMs achieving approximately 90% compatibility with human tumor board recommendations in standard cases, but expert judgment remains indispensable, particularly for rare cancers, edge cases, and decisions requiring patient context (comorbidities, prior toxicities, preferences, social determinants, resource availability). Roche’s implementation demonstrates the appropriate model: LLMs match patients with NCCN guidelines and “ensure each patient receives tailored treatment recommendations,” but oncologists make final management decisions after What infrastructure is required to support medical LLM integration in tumor boards?
MiBA’s infrastructure processing 1.4 million physician notes and approximately 1 million PDF reports demonstrates requirements: data ingestion from multiple EHR sources (clinical notes, pathology reports, radiology findings, genetic results), scalable NLP processing for entity extraction and relationship mapping, Medical LLM for clinical reasoning and recommendation generation, retrieval-augmented generation systems for guideline currency, and governance frameworks for audit trails, human review workflows, and provenance tracking. Organizations need: document processing pipelines, entity extraction and normalization capabilities, secure storage for patient data, and integration with tumor board documentation systems.
How do organizations validate medical LLM recommendations for clinical use?
Roche’s approach demonstrates validation pathways: compare LLM recommendations against NCCN guideline-concordant options for alignment with evidence standards. MiBA’s methodology establishes accuracy benchmarks through comparison with manually reviewed truth sets, achieving 93% entity extraction and 88% relationship extraction F1-scores. Organizations implementing regulatory-grade oncology data curation emphasize “uncompromising quality” through human-in-the-loop workflows where oncologists validate automated outputs before clinical use. Prospective pilots introduce LLM suggestions alongside traditional preparation, with tumor boards assessing recommendation quality, clinical utility, and evidence appropriateness before operational deployment.
What are the main limitations of current medical LLMs for tumor board decision support?
Performance variability on rare cancers remains a challenge, sarcoma tumor board evaluation found “moderate effectiveness” compared to higher accuracy on common tumor types. Studies note that LLMs occasionally suggest therapies based on weaker evidence not reflecting institutional protocols. Multimodal integration remains incomplete, current systems process textual reports rather than directly analyzing imaging or pathology slides. Most importantly, prospective trials measuring patient outcomes (response rates, survival, quality of life) for LLM-supported versus traditional tumor boards are not yet available. The systematic review of 56 studies across 15 cancer types represents substantial evidence, but longitudinal outcome validation is still emerging.
How do medical LLMs ensure recommendations stay current with evolving oncology guidelines?
Retrieval-augmented generation (RAG) architectures address guideline currency by dynamically fetching up-to-date evidence during recommendation generation rather than relying solely on pre-trained knowledge. The MEREDITH system combines LLM reasoning with retrieval over clinical evidence and molecular data, ensuring recommendations incorporate current guidelines, recently approved therapies, and active clinical trials. Roche’s NCCN guideline matching demonstrates this approach in practice, the system aligns patient profiles with “the most relevant clinical guidelines” by querying current guideline databases rather than depending on static training data. Organizations must implement update processes to refresh guideline repositories as NCCN and institutional protocols evolve.