When Guidelines Central partnered with John Snow Labs to match patients with clinical guidelines from 35+ medical societies, the technical challenge was not generating recommendations. Large language models can produce fluent clinical suggestions. The challenge was ensuring those recommendations were factually accurate, mapped to the correct guideline section, and delivered in structured form that electronic health record vendors could integrate.
The gap between “fluent text” and “actionable clinical decision” defines why healthcare organizations cannot deploy large language models alone for clinical decision support. General-purpose models generate unstructured suggestions. Clinical decision support systems require structured, validated, coded data that integrates with workflows, triggers alerts, and feeds registries. Between the two lies the “last mile” problem: transforming narrative output into machine-readable decisions that clinicians can trust.
The precision gap that general-purpose models cannot close
The Department of Veterans Affairs discovered this limitation when piloting large language model interfaces for clinical data discovery across their health system serving over 9 million veterans. Out-of-the-box accuracy of current large language models on clinical notes was unacceptable. However, as R. Spencer Schaefer, Chief Health Informatics Officer at Kansas City VAMC recently explained, accuracy could be significantly improved with pre-processing: using clinical text summarization models prior to feeding content to generative artificial intelligence significantly improved output quality.
This finding validates systematic benchmarking that demonstrates why specialized healthcare natural language processing outperforms general-purpose models on clinical text. Assessment of 48 medical expert-annotated clinical documents showed Healthcare NLP achieving 96% F1-score for protected health information detection, significantly outperforming GPT-4o at 79%. More critically, GPT-4o completely missed 14.6% of entities while Healthcare NLP missed only 0.9%. For clinical decision support, a 14.6% complete miss rate means critical information never enters decision logic.
The CLEVER study’s blind physician evaluation reinforced this performance gap across clinical tasks. Medical doctors preferred specialized healthcare natural language processing 45% to 92% more often than GPT-4o on factuality, clinical relevance, and conciseness across 500 novel clinical test cases. When physicians evaluating clinical summarization preferred specialized models 48% to 25% on clinical relevance, the message is clear: fluency does not equal clinical utility.
Why unstructured LLM output breaks clinical workflows
Robust Clinical Decision Support Systems (CDSS) cannot consume narrative text. They require structured, coded data that can match against clinical guidelines, trigger alerts, feed dashboards, and integrate into electronic health record workflows. Large language model outputs fail on multiple dimensions:
Missing clinical context: A model might generate “the patient has pneumonia” when the clinical note actually states “consider pneumonia,” representing a physician’s diagnostic hypothesis. Without assertion detection and negation handling, downstream systems risk misclassifying a potential condition as a confirmed diagnosis. Healthcare NLP processes over 2,800 pre-trained clinical models for entity extraction, relation extraction, and assertion detection specifically to capture this context.
No standardized terminology: Large language models use variable phrasing. Clinical decision support requires normalization to standard medical terminologies including SNOMED CT, ICD-10, LOINC, and RxNorm. Without this mapping, decision logic cannot match extracted findings against guideline criteria or registry definitions.
Inability to integrate with existing systems: Electronic health records, quality measure calculators, and registry reporting tools expect structured, coded inputs. Narrative text requires human interpretation before it can feed these workflows.
vCare Companion’s implementation, as its CEO Jonathan McCoy explains, demonstrates the operational requirement. Their robot helps hospital staff reduce administrative burden by over 3 hours per shift through ambient listening to patient-staff conversations, automatically filling medical forms, and integrating with electronic medical records at point of care. This works because John Snow Labs’ Medical Language Models perform clinical information extraction and normalization, not just transcription. The system extracts structured clinical entities that populate discrete fields in medical forms, enabling direct electronic health record integration.
How production systems structure LLM output for clinical use
Vickie Reyes, Director of Informatics and Clinical Decision Support at Guideline Central, presented a session in AI Summit 2025 that perfectly illustrates a complete solution architecture pipeline. The Guideline Central system answers detailed questions about clinical guideline documents, matches patient cases presented as unstructured text to the right guideline section, and provides deep links with explanations. This requires healthcare-specific large language models tuned for reasoning on clinical guidelines, combined with structured extraction that identifies patient conditions, procedural history, and clinical context in machine-readable form.
The architecture is available as both a standalone tool for clinicians and an embeddable module for electronic health record and clinical decision support vendors, specifically because it outputs structured, actionable recommendations rather than unstructured suggestions.
Another example of these kinds of systems in oncology was described by Vishakha Sharma, Senior Principal Data Scientist at Roche. Roche’s system applies healthcare-specific large language models to construct detailed oncology patient timelines from diverse clinical notes: primary care providers, oncologists, discharge summaries, emergency departments, pathology, and radiology reports. More importantly, it matches patients with National Comprehensive Cancer Network clinical guidelines by analyzing comprehensive patient data including genetic, epigenetic, and phenotypic information, then accurately aligning individual patient profiles with relevant guidelines. This enhances precision in oncology care by ensuring each patient receives tailored treatment recommendations.
The critical architectural component is not the large language model alone. It is the natural language processing pipeline that extracts structured clinical entities, maps them to standard terminologies, detects temporal relationships and clinical context, and outputs FHIR-compliant or tabular data that downstream decision systems can process.
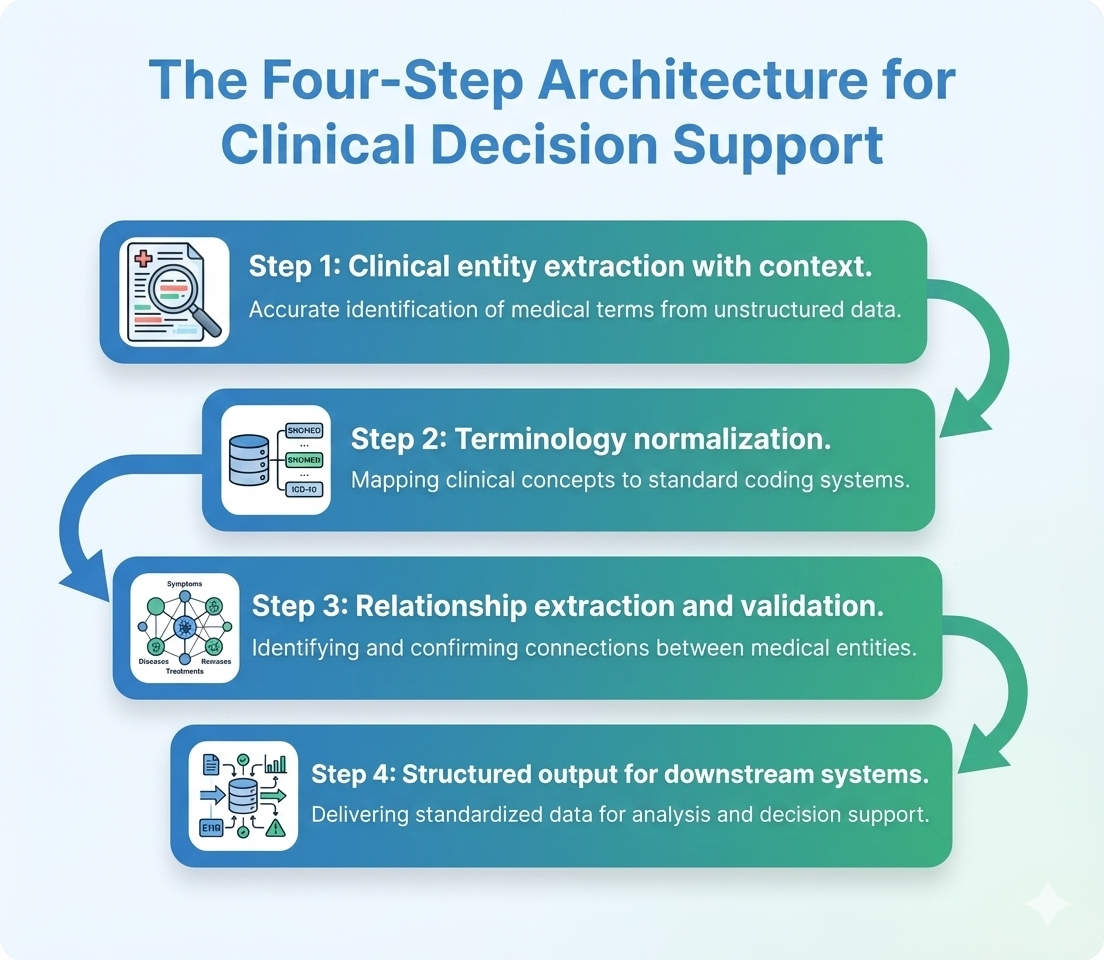
The four-step architecture from text to action
Across the implementations we have seen that successfully deploy generative artificial intelligence for clinical decision support, four processing steps emerge as essential:
Step 1: Clinical entity extraction with context Extract diagnoses, medications, procedures, laboratory values, and measurements while capturing negation, temporality, certainty, and anatomical context. This distinguishes “patient has diabetes” from “patient denies diabetes” or “family history of diabetes.”
Step 2: Terminology normalization Map extracted entities to standard medical vocabularies. “Blood sugar” becomes LOINC code 2345-7 for glucose. “Heart attack” becomes ICD-10 code I21 for acute myocardial infarction. This standardization enables matching against guideline criteria and quality measures.
Step 3: Relationship extraction and validation Identify clinical relationships: which medication treats which condition, which laboratory value indicates which disease state, which procedure was performed for which indication. Validate extracted data against known patient information to catch inconsistencies.
Step 4: Structured output for downstream systems Format validated, coded data for clinical decision support engines, quality measure calculators, registry reporting, or billing systems. The output must be machine-readable and workflow-integrated, not requiring human interpretation.
Why specialized models outperform general-purpose systems
The CLEVER study provides the definitive answer. Superior performance was achieved using an 8-billion-parameter model, demonstrating that domain-specific training enables smaller models to outperform much larger general-purpose systems in precision-critical tasks. This has practical implications: specialized healthcare natural language processing operates with fixed-cost local deployment rather than per-request cloud API pricing. Healthcare NLP reduces processing costs by over 80% compared to Azure and GPT-4o.
Precision and cost efficiency stem from the same architectural decision: building models specifically for healthcare rather than adapting general-purpose systems. Organizations can explore Healthcare NLP capabilities, review Medical LLM implementations, or access live demonstrations to see extraction and structuring performance on clinical text.
From fluent text to trusted decisions
The implementations across Guidelines Central, Roche, vCare Companion, and VA demonstrate a consistent pattern: generative artificial intelligence produces valuable clinical insights, but specialized natural language processing makes those insights actionable. The gap between fluent output and clinical decision support is not cosmetic. It is the difference between text a human must interpret and structured data that downstream systems can process, validate, and integrate into workflows.
Healthcare organizations deploying large language models for clinical applications must architect the complete pipeline: generation combined with extraction, normalization, validation, and structured output. The technology exists. The accuracy has been validated. The production deployments are operational. The question is whether organizations will build closed-loop systems that transform generative artificial intelligence from suggestion engine into decision support, or whether they will deploy fluent text generators and wonder why clinical workflows remain unchanged.
FAQs
Why cannot large language models alone be used for clinical decision support?
Large language models generate unstructured narrative text. Clinical decision support systems require structured, coded data that can match against clinical guidelines, trigger alerts, and integrate with electronic health record workflows. Without natural language processing pipelines that extract entities, detect clinical context (negation, temporality, certainty), and normalize to standard terminologies like SNOMED CT and ICD-10, large language model outputs cannot feed decision logic. Out-of-the-box accuracy of current large language models on clinical notes is usually insufficient, but can be significantly improved using clinical text summarization models for pre-processing. This is why specialized Healthcare NLP is essential between generative artificial intelligence and clinical workflows.
What is the accuracy difference between specialized healthcare NLP and general-purpose LLMs on clinical text?
Systematic assessment on 48 medical expert-annotated clinical documents showed Healthcare NLP achieving 96% F1-score for protected health information detection compared to GPT-4o’s 79%. More critically, GPT-4o completely missed 14.6% of entities while Healthcare NLP missed only 0.9%. The CLEVER study‘s blind physician evaluation found medical doctors preferred specialized healthcare natural language processing 45% to 92% more often than GPT-4o on factuality, clinical relevance, and conciseness across 500 novel clinical test cases. For clinical decision support where missed entities break decision logic, this 17-percentage-point precision gap defines which systems can be trusted for production deployment.
How does clinical NLP handle negation and context that LLMs often miss?
Healthcare NLP uses over 2,800 pre-trained clinical models for entity extraction, relation extraction, and assertion detection. Assertion detection specifically identifies whether a condition is present, absent, hypothetical, historical, or associated with someone other than the patient. This distinguishes “patient has diabetes” from “patient denies diabetes” or “family history of diabetes.”
What does an end-to-end pipeline look like from LLM output to clinical decision?
Guidelines Central’s architecture shows the complete flow: healthcare-specific large language models answer questions about clinical guidelines and match patient cases to relevant guideline sections, combined with structured extraction that outputs machine-readable recommendations. The system is available as an embeddable module for electronic health record vendors specifically because it produces structured output, not just narrative text. A typical pipeline includes: (1) Large language model generates clinical summary or recommendation, (2) Natural language processing extracts entities with context, (3) Terminology normalization maps to SNOMED CT, ICD-10, LOINC, (4) Relationship extraction identifies clinical connections, (5) Validation checks against patient data, (6) Structured output feeds clinical decision support, quality measures, or registry reporting. Organizations can review Generative AI Lab for human-in-the-loop validation workflows.
What is the cost difference between specialized healthcare NLP and cloud-based LLM APIs for clinical text processing?
Healthcare NLP operates with fixed-cost local deployment rather than per-request cloud API pricing. Systematic benchmarking found Healthcare NLP reduces processing costs by over 80% compared to Azure and GPT-4o when processing clinical documents at scale. This cost advantage stems from the same architectural decision that improves accuracy: an 8-billion-parameter model trained specifically for healthcare can be deployed on-premise, avoiding per-request charges while outperforming much larger general-purpose systems. For organizations processing millions of clinical notes annually, the cost differential is substantial. Additionally, on-premise deployment addresses data privacy requirements and eliminates dependency on external API availability. Organizations can explore deployment options and integration patterns for Healthcare NLP and Medical LLM across Databricks, AWS, Azure, and on-premise environments.