We used John Snow Labs’ Healthcare NLP & LLM library to train a custom Named Entity Recognition (NER) model that automatically extracts key medical entities from clinical text, including vaccine types, infectious diseases, other diseases, and symptoms. This end-to-end NLP pipeline enables scalable, accurate information extraction, which is critical for public health monitoring, pharmacovigilance, and research on vaccine-related outcomes. It significantly reduces manual effort and paves the way for a faster, data-driven decision-making in healthcare.
Introduction
Infectious diseases are illnesses caused by microscopic organisms such as bacteria, viruses, fungi, or parasites that invade the body and multiply. These microorganisms are found everywhere in nature — water, soil, plants, and animals. While many are harmless or even beneficial, some can cause diseases under certain conditions. Infectious diseases can spread from person to person, through contaminated food or water, by insects or animals, or via environmental exposure.
Globally, infectious diseases remain a leading cause of illness and death, especially among young children and vulnerable populations. Lower respiratory infections, diarrheal diseases, malaria, tuberculosis, and measles are among the top causes of mortality worldwide, with many of these being preventable through public health measures and vaccination.
Vaccines are one of the most effective tools in modern medicine for preventing infectious diseases. They work by training the immune system to recognize and fight specific pathogens without causing the disease itself. This process not only protects vaccinated individuals but also contributes to “herd immunity,” reducing the overall spread of disease and protecting those who cannot be vaccinated due to medical reasons.
Vaccination has led to the elimination or dramatic reduction of diseases such as smallpox, polio, measles, and diphtheria in many parts of the world. Immunization currently prevents 3.5 to 5 million deaths every year from diseases like diphtheria, tetanus, pertussis (whooping cough), influenza, and measles. However, lapses in vaccination coverage can lead to outbreaks and the resurgence of diseases previously under control.
Vaccination provide the power to control, eliminate, and even eradicate some of the world’s most dangerous infectious diseases, saving millions of lives each year.
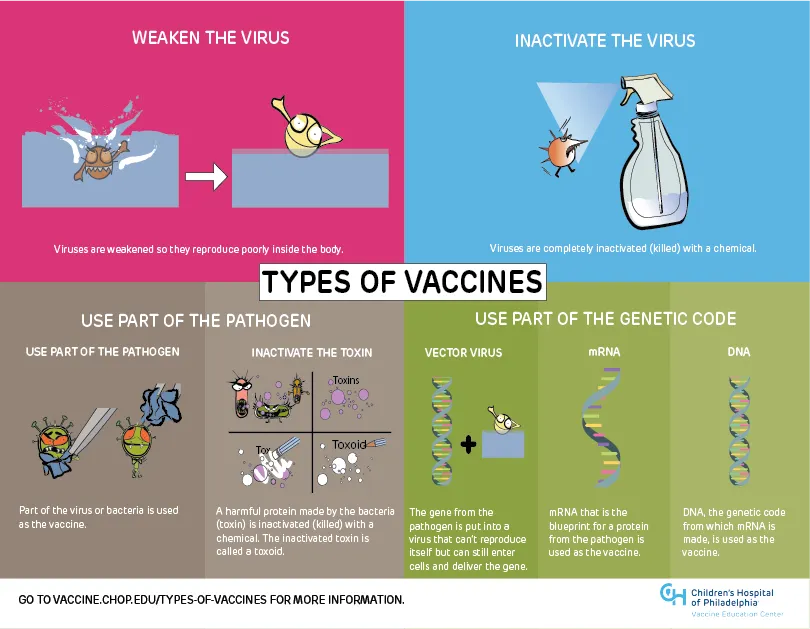
Vaccine- and Disease-related Infographics, Children’s Hospital of Philadelphia
Extracting Entities from Clinical/Medical Text
NER models are essential for identifying and categorizing entities within text. In the context of vaccines and infectious diseases analysis, our model can automatically recognize mentions of various vaccines, infectious diseases, other diseases, signs and symptoms, and other key entities (a total of 14 entities) from large volumes of unstructured clinical or biomedical documentation.
Here are the entities extracted by this model:
- Bacterial_Vax: A vaccine designed to protect against bacterial infections (e.g., pneumococcal or meningococcal vaccines).
- Viral_Vax: A vaccine developed to prevent viral infections such as influenza, hepatitis, or COVID-19.
- Cancer_Vax: A therapeutic or preventive vaccine aimed at stimulating the immune system to target cancer cells.
- Bac_Vir_Comb: A combination vaccine that provides protection against both bacterial and viral pathogens.
- Other_Vax: Vaccines characterized by their components rather than the target pathogen. This category includes vaccines composed of polysaccharides, proteins, subunits, conjugates, or other non-whole-pathogen elements, which do not neatly fall under bacterial, viral, or cancer vaccine types.
- Vax_Dose: Information indicating the amount, number, or schedule of vaccine doses.
- Infectious_Disease: A disease caused by pathogenic microorganisms such as bacteria, viruses, or fungi that can spread directly or indirectly.
- Other_Disease_Disorder: A non-infectious or unrelated medical condition or disorder mentioned in the text.
- Sign_Symptom: Observable signs or reported symptoms that may indicate the presence of a disease or reaction.
- Toxoid: A modified bacterial toxin used as a vaccine component to elicit immunity without causing disease (e.g., tetanus toxoid).
- Adaptive_Immunity: A reference to the immune system’s specific response to antigens through T-cells and B-cells, often induced by vaccination.
- Inactivated: A vaccine composed of pathogens that have been killed or rendered non-infectious while still triggering an immune response.
- Date: Any calendar reference related to vaccination events, symptoms onset, or medical history.
- Age: A specific mention of a person’s age, age group.
Pretrained pipelines in the Healthcare NLP library make it possible to uncover and organize valuable insights from unstructured text, converting them into structured datasets that help deeper analysis and informed decision-making.
In this case, we use one line of code to process the pretrained pipeline (vaccine_names), which is specifically trained to extract vaccines, diseases and certain other entities:
pipeline = PretrainedPipeline("vaccine_names", "en", "clinical/models")
result = pipeline.fullAnnotate(text)
Extracting entities in a structured format improves usability and integration by enabling efficient retrieval and detailed analysis of patient information. It ensures consistency and standardization, which are essential for advanced analytics and accurate decision-making. By converting unstructured text into a structured format, this approach delivers actionable insights that enhance patient care, support impactful research, and guide public health strategies.
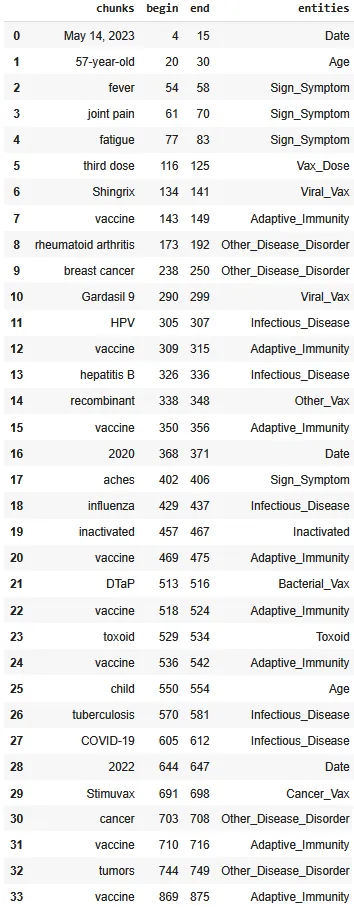
Dataframe providing the chunks, and the assigned labels.
The ability to quickly visualize the entities is a very useful feature for examining the generated results. Spark NLP Display is an open-source Python library for visualizing the extracted and labeled entities. NerVisualizer highlights the extracted named entities and also displays their labels as decorations on top of the analyzed text.
This tool simplifies the process of understanding and interpreting extracted data, aiding in model validation, pattern recognition, and insight generation from unstructured medical text. By enhancing data analysis and interpretation, the visualizer supports more informed decision-making in healthcare.
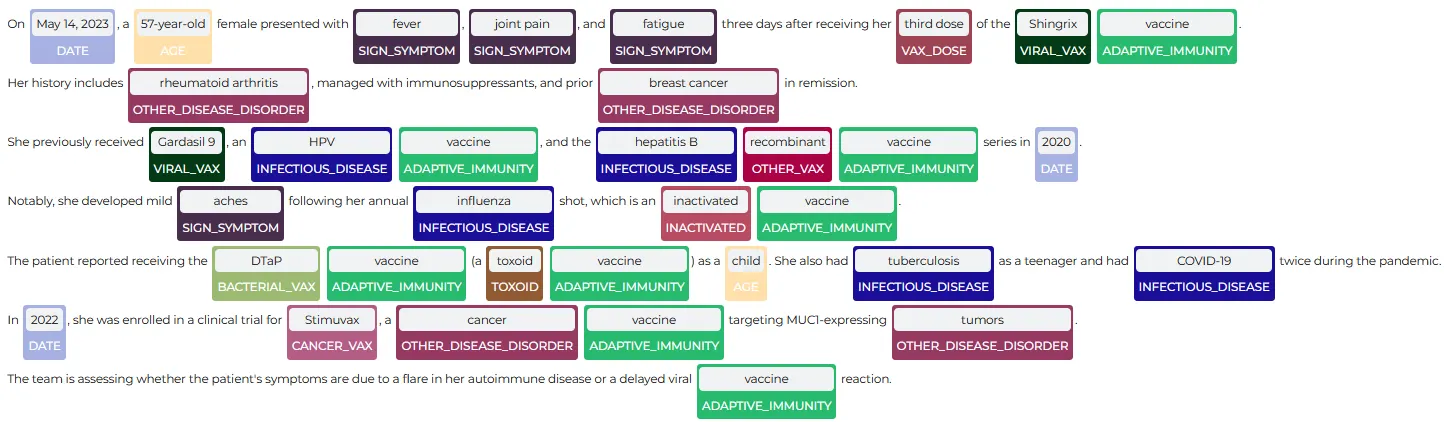
The NerVisualizer highlights the named entities that are identified by the model and also displays their labels as decorations on top of the analyzed text.
The results demonstrate the model’s effectiveness in accurately identifying and categorizing vaccine-related entities from clinical text. These findings validate the utility of the NLP pipeline in real-world scenarios, highlighting its potential for accelerating information extraction at scale.
Conclusion
In this work, we demonstrated how a custom NER model, built using John Snow Labs’ Healthcare NLP & LLM library, can effectively extract vaccine-related information from unstructured clinical text. By identifying entities such as vaccine types, infectious and other diseases, symptoms, and immunization details, the pipeline provides a structured format that supports more efficient data analysis and decision-making. The ability to automate this extraction at scale offers clear benefits for clinical research, epidemiological studies, and vaccine safety monitoring.
These results highlight the practical value of applying domain-specific NLP to extract structured vaccine-related data from real-world text. By enabling faster, more consistent access to critical clinical information, the model can support a wide range of applications, from improving patient care workflows to enhancing vaccine research and public health reporting. As healthcare data continues to grow in volume and complexity, tools like this NER pipeline will play an increasingly important role in turning unstructured information into meaningful, actionable insights.
John Snow Labs and Medical Language Models
John Snow Labs, offers a powerful NLP & LLM library tailored for healthcare, empowering professionals to extract actionable insights from medical text. Utilizing advanced AI techniques like Named Entity Recognition (NER), assertion status detection, relation extraction, Question-Answering, and summarizing, this library helps uncover vital genetics information for more accurate diagnosis, treatment, and prevention.
Medical Language Models
The Healthcare Library is a powerful component of John Snow Labs’ Healthcare NLP platform, designed to facilitate NLP tasks within the healthcare domain. This library provides over 2,700 pre-trained models and pipelines tailored for medical data, enabling accurate information extraction, NER for clinical and medical concepts, and text analysis capabilities. Regularly updated and built with cutting-edge algorithms, the Healthcare library aims to streamline information processing and empower healthcare professionals with deeper insights from unstructured medical data sources, such as electronic health records, clinical notes, and biomedical literature.
John Snow Labs has created custom large language models (LLMs) tailored for diverse healthcare use cases. These models come in different sizes and quantization levels, designed to handle tasks such as summarizing medical notes, answering questions, performing retrieval-augmented generation (RAG), named entity recognition and facilitating healthcare-related chats.
John Snow Labs’ GitHub repository serves as a collaborative platform where users can access open-source resources, including code samples, tutorials, and projects, to further enhance their understanding and utilization of Healthcare NLP and related tools.
John Snow Labs also offers periodic certification training to help users gain expertise in utilizing the Healthcare Library and other components of their NLP platform.
John Snow Labs’ demo page provides a user-friendly interface for exploring the capabilities of the library, allowing users to interactively test and visualize various functionalities and models, facilitating a deeper understanding of how these tools can be applied to real-world scenarios in healthcare and other domains.
This project has been funded in whole or in part with Federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract №75N93024C00010.