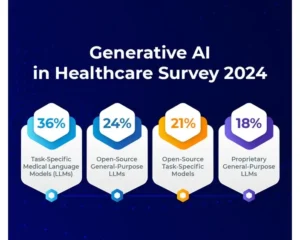
Pretrained AI pipelines streamline complex processes into a single line of code, offering easy access, superior performance, resource savings, smooth integration, and continuous updates.
In this blog post, we’ll see that instead of creating long pipelines, we can achieve the same result with pre-trained pipelines. Let’s start with a brief introduction to Spark NLP and then discuss the details of pretrained pipelines with some concrete results.
Spark NLP & LLM
The Healthcare Library is a powerful component of John Snow Labs’ Spark NLP platform, designed to facilitate NLP tasks within the healthcare domain. This library provides over 2,200 pre-trained models and pipelines tailored for medical data, enabling accurate information extraction, NER for clinical and medical concepts, and text analysis capabilities. Regularly updated and built with cutting-edge algorithms, the Healthcare library aims to streamline information processing and empower healthcare professionals with deeper insights from unstructured medical data sources, such as electronic health records, clinical notes, and biomedical literature.
John Snow Labs’ GitHub repository serves as a collaborative platform where users can access open-source resources, including code samples, tutorials, and projects, to further enhance their understanding and utilization of Spark NLP and related tools.
John Snow Labs also offers periodic certification training to help users gain expertise in utilizing the Healthcare Library and other components of their NLP platform.
John Snow Labs’ demo page provides a user-friendly interface for exploring the capabilities of the library, allowing users to interactively test and visualize various functionalities and models, facilitating a deeper understanding of how these tools can be applied to real-world scenarios in healthcare and other domains.
Introduction to Spark NLP
Spark NLP processes the data using Pipelines, the structure that contains all the steps to be run on the input data:
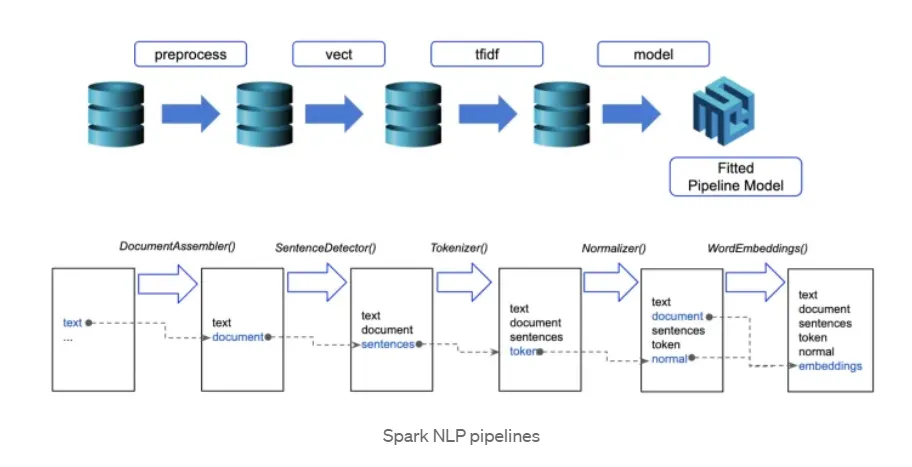
Each step contains an annotator that performs a specific task such as tokenization, normalization, and dependency parsing. Each annotator has input(s) annotation(s) and outputs new annotation.
An annotator in Spark NLP is a component that performs a specific NLP task on a text document and adds annotations to it. An annotator takes an input text document and produces an output document with additional metadata, which can be used for further processing or analysis. For example, a named entity recognizer annotator might identify and tag entities such as people, organizations, and locations in a text document, while a sentiment analysis annotator might classify the sentiment of the text as positive, negative, or neutral.
Pre-trained Pipelines
Managing health records manually can be very complex and time-consuming to fully understand. Therefore, automating this process can be very useful. Traditionally manual, this process can now be partially or fully automated using Natural Language Processing (NLP) techniques. This allows for faster and more efficient analysis of health records, which is a huge advantage for healthcare providers.
A Pipeline is a concept used in machine learning to manipulate and learn from data by applying different algorithms. Especially in Apache Spark ML, this process performs a series of operations in a specific order and facilitates data flow while passing the results of these operations to the next step.
For example, executing a workflow in a Pipeline might include the following steps in a text document:
– Splitting each document’s text into sentences and tokens (words).
– Converting each token into a numerical feature vector (e.g. word embeddings).
– Using a NER model to identify and classify named entities within text data, facilitating information extraction and downstream NLP tasks.
Each step in the NLP pipeline uses different Transformers and Estimators, all smoothly working together to create a complete process. This coordinated effort automates and manages the complexities of artificial intelligence tasks effectively, promoting seamless cooperation in data processing and learning activities.
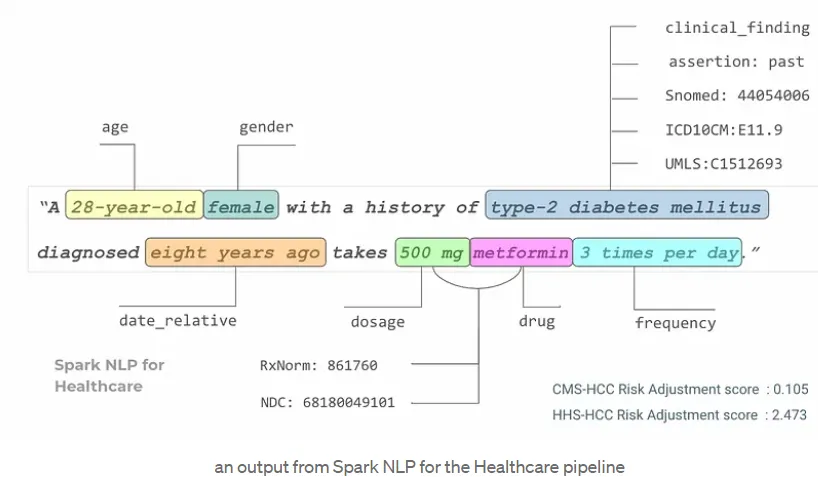
With a Healthcare NLP pipeline, it is possible to perform multiple tasks on clinical text in a single workflow: extract relevant clinical information, determine whether that information is present, hypothetical, or absent (assertion status), and map the extracted concepts to standardized medical codes like ICD, RxNorm, or SNOMED CT codes.
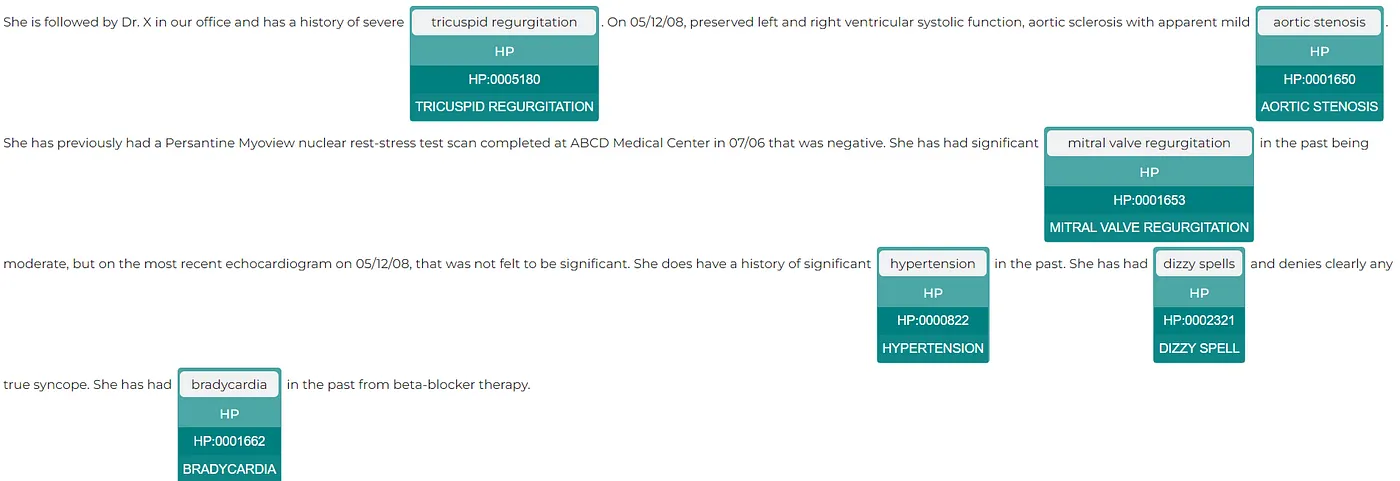
As an example, the pipeline below is used for mapping relevant medical concepts from unstructured text data to standardized medical coding systems. The pipeline first detects (first six stages) mentions of genes (labels them as GENE) and human phenotypes (HP) in a medical text and then assigns the Human Phenotype Ontology codes, which are a structured and controlled vocabulary for describing phenotypic abnormalities in human beings.
document_assembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentence_detector = SentenceDetectorDLModel.pretrained("sentence_detector_dl_healthcare", "en", "clinical/models") \
.setInputCols(["document"])\
.setOutputCol("sentence")
tokenizer = Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")
word_embeddings = WordEmbeddingsModel.pretrained("embeddings_clinical", "en", "clinical/models")\
.setInputCols(["sentence", "token"])\
.setOutputCol("word_embeddings")
ner = MedicalNerModel.pretrained("ner_human_phenotype_gene_clinical", "en", "clinical/models") \
.setInputCols(["sentence", "token", "word_embeddings"]) \
.setOutputCol("ner")\
ner_converter = NerConverterInternal()\
.setInputCols(["sentence", "token", "ner"])\
.setOutputCol("ner_chunk")\
.setWhiteList(["HP"])
chunk2doc = Chunk2Doc()\
.setInputCols("ner_chunk")\
.setOutputCol("ner_chunk_doc")
sbert_embedder = BertSentenceEmbeddings.pretrained("sbiobert_base_cased_mli",'en','clinical/models')\
.setInputCols(["ner_chunk_doc"])\
.setOutputCol("sbert_embeddings")\
.setCaseSensitive(False)
resolver = SentenceEntityResolverModel.pretrained("sbiobertresolve_HPO", "en", "clinical/models") \
.setInputCols(["sbert_embeddings"]) \
.setOutputCol("resolution")\
.setDistanceFunction("EUCLIDEAN")
pipeline = Pipeline(stages = [
document_assembler,
sentence_detector,
tokenizer,
word_embeddings,
ner,
ner_converter,
chunk2doc,
sbert_embedder,
resolver])
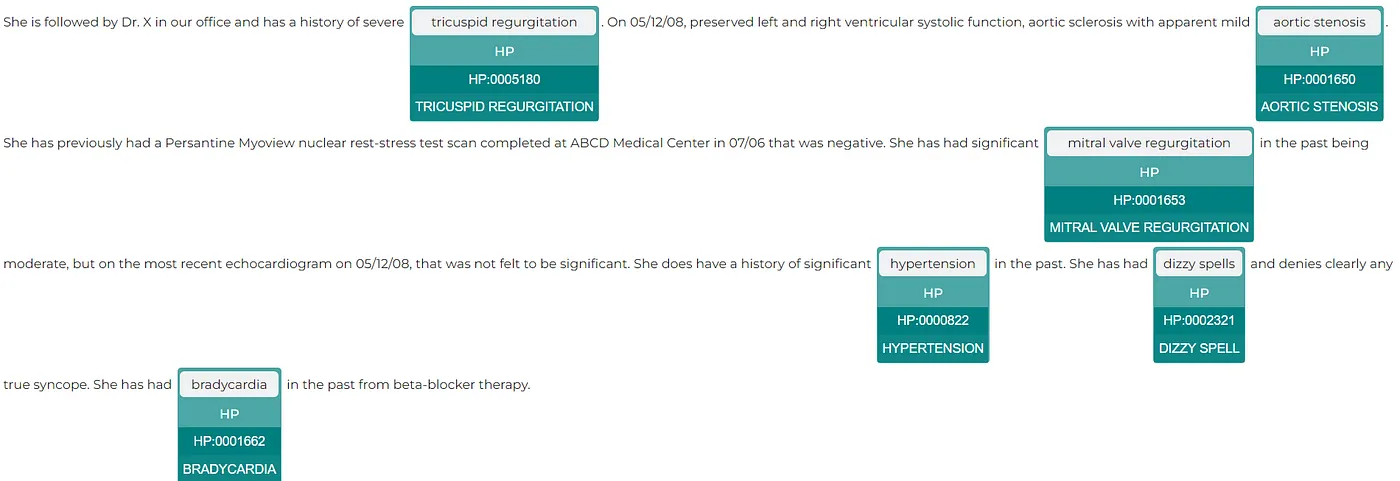
text = """She is followed by Dr. X in our office and has a history of severe tricuspid regurgitation. On 05/12/08, preserved left and right ventricular systolic function, aortic sclerosis with apparent mild aortic stenosis. She has previously had a Persantine Myoview nuclear rest-stress test scan completed at ABCD Medical Center in 07/06 that was negative. She has had significant mitral valve regurgitation in the past being moderate, but on the most recent echocardiogram on 05/12/08, that was not felt to be significant. She does have a history of significant hypertension in the past. She has had dizzy spells and denies clearly any true syncope. She has had bradycardia in the past from beta-blocker therapy."""
data = spark.createDataFrame([[text]]).toDF("text")
result = pipeline.fit(data).transform(data)
The ability to quickly visualize the entities generated using Spark NLP is a very useful feature for speeding up the development process as well as for understanding the obtained results. Spark NLP Display is an open-source Python library for visualizing the annotations generated with Spark NLP.
The EntityResolverVisualizer annotator highlights the extracted named entities, displaying their labels and the corresponding HPO resolver codes. The colors assigned to the predicted labels can be configured to fit the particular needs of the application.
Creating a pipeline can sometimes be a complex process. Determining which models or parameters to use can be difficult and time-consuming. Therefore, to simplify things, these long pipelines were trained as pre-trained pipelines in just one line of code. Thus, instead of writing a long pipeline, a single line of pre-trained pipelines with the same functionality will be sufficient. For an extended example of usage, see the pre-trained pipelines notebook in the Spark NLP Workshop repository.
Using a pre-trained pipeline provided a quick and efficient way to produce the same results, saving time and effort compared to developing a complete pipeline from scratch.
from sparknlp.pretrained import PretrainedPipeline
ner_pipeline = PretrainedPipeline("hpo_resolver_pipeline", "en", "clinical/models")
result = ner_pipeline.annotate("""She is followed by Dr. X in our office and has a history of severe tricuspid regurgitation. On 05/12/08, preserved left and right ventricular systolic function, aortic sclerosis with apparent mild aortic stenosis. She has previously had a Persantine Myoview nuclear rest-stress test scan completed at ABCD Medical Center in 07/06 that was negative. She has had significant mitral valve regurgitation in the past being moderate, but on the most recent echocardiogram on 05/12/08, that was not felt to be significant. She does have a history of significant hypertension in the past. She has had dizzy spells and denies clearly any true syncope. She has had bradycardia in the past from beta-blocker therapy.""")
Here is the list of Task-Based Clinical Pretrained Pipelines offered by Spark NLP.
| index | pipeline | description |
|---|---|---|
| 1 | explain_clinical_doc_generic | Extract clinical/medical entities at a generic level, assign assertion status, and establish relations between extracted entities. |
| 2 | explain_clinical_doc_granular | Extract clinical/medical entities at a granular level, assign assertion status, and establish relations between extracted entities. |
| 3 | explain_clinical_doc_biomarker | Extract biomarkers and their results, classify sentences based on whether they include a biomarker mention or not, and pick up relations between biomarkers and their results. |
| 4 | explain_clinical_doc_oncology | Extract cancer-related information from clinical documents, assign assertion status, and provide relations between the extracted data. |
| 5 | explain_clinical_doc_radiology | Extract radiology entities, assign assertion status, and find relationships between the entities. |
| 6 | explain_clinical_doc_vop | Extract healthcare-related entities, assign assertion status, and relate extracted entities from the voice of patients in documents. |
| 7 | explain_clinical_doc_carp | Extract clinical and medication entities, assign assertion status to the clinical items, and establish relationships between those entities. |
| 8 | explain_clinical_doc_era | Extract clinical events, assign assertion status, and designate temporal relationships between them. |
| 9 | explain_clinical_doc_ade | Extract ADE and DRUG entities from text, classify if the text involves ADEs, assign assertion status to ADE entities, and relate Drugs with their ADEs. |
| 10 | explain_clinical_doc_medication | Extract medication entities, assign assertion status to the extracted items, and establish relationships between those entities. |
| 11 | explain_clinical_doc_risk_factors | Extract all clinical/medical entities, which may be considered as risk factors , assign assertion status to the entities, provide relations between the entities |
| 12 | explain_clinical_doc_public_health | Extract public health-related entities, assign assertion status to the entities, establish relations between the extracted entities . |
Using those pre-trained pipelines provides a lot of benefits. These pipelines, crafted by experts, relieve the need for extensive model training or selecting the best models out of thousands of alternatives in the Spark NLP Healthcare Library, saving both time and resources. Their optimized design ensures superior performance, especially valuable for users with limited data or expertise. Additionally, pre-trained pipelines are user-friendly and seamlessly integrated. Furthermore, regular, and frequent updates and maintenance by developers ensure that these pipelines stay at the forefront of NLP advancements.
Let’s take the explain_clinical_doc granular pre-trained pipeline as an example.
You can download the pre-trained pipeline from model hubs like this:
from sparknlp.pretrained import PretrainedPipeline
pipeline = PretrainedPipeline("explain_clinical_doc_granular", "en", "clinical/models")
Once you’ve downloaded the pre-trained pipeline, accessing and utilizing it is simple with the .from_disk method. This method seamlessly loads the pre-trained pipeline directly from your local storage.
from sparknlp.pretrained import PretrainedPipeline
pipeline = PretrainedPipeline.from_disk("/root/cache_pretrained/explain_clinical_doc_granular_en_5.2.1_3.0_1706288377782")
Let’s use LightPipeline here to extract the entities. LightPipeline is a Spark NLP specific Pipeline class equivalent to the Spark ML Pipeline. The difference is that its execution does not hold to Spark principles, instead, it computes everything locally (but in parallel) in order to achieve fast results when dealing with small amounts of data.
To retrieve detailed information on annotations, we can use fullAnnotate() to return a dictionary list of the entire annotations’ content.
text ="""The patient admitted for gastrointestinal pathology, under working treatment. History of prior heart murmur with echocardiogram findings as above on March 1998. According to the latest echocardiogram, basically revealed normal left ventricular function with left atrial enlargement . Based on the above findings, we will treat her medically with ACE inhibitors and diuretics and see how she fares. """ result = pipeline.fullAnnotate(text)[0]
NER Result:
The provided code allows us to generate a data frame that combines the extracted text segments (chunks) with the corresponding labels assigned by the NER model (entities or labels).
import pandas as pd
chunks=[]
entities=[]
begins=[]
ends=[]
for n in result['jsl_ner_chunk']:
chunks.append(n.result)
begins.append(n.begin)
ends.append(n.end)
entities.append(n.metadata['entity'])
df = pd.DataFrame({'chunks':chunks, 'begin':begins, 'end':ends, 'entities':entities})
df
| | chunks | begin | end | entities | |---:|:---------------------------|--------:|------:|:--------------------| | 0 | admitted | 12 | 19 | Admission_Discharge | | 1 | gastrointestinal pathology | 25 | 50 | Clinical_Dept | | 2 | heart murmur | 95 | 106 | Heart_Disease | | 3 | echocardiogram | 113 | 126 | Test | | 4 | March 1998 | 149 | 158 | Date | | 5 | echocardiogram | 185 | 198 | Test | | 6 | normal | 220 | 225 | Test_Result | | 7 | left ventricular function | 227 | 251 | Test | | 8 | left atrial enlargement | 258 | 280 | Heart_Disease | | 9 | her | 327 | 329 | Gender | | 10 | ACE inhibitors | 346 | 359 | Drug_Ingredient | | 11 | diuretics | 365 | 373 | Drug_Ingredient | | 12 | she | 387 | 389 | Gender |
Assertion Status Result:
The pipeline also assigns assertion status to clinical entities such as Present, Absent, Hypothetical, etc, extracted by NER based on their context in the text.
chunks=[]
entities=[]
status=[]
for n,m in zip(result['assertion_ner_chunk'],result['assertion']):
chunks.append(n.result)
entities.append(n.metadata['entity'])
status.append(m.result)
df = pd.DataFrame({'chunks':chunks, 'entities':entities, 'assertion':status})
df
|sentence_id|begin|end|entity |label |assertion_status| +-----------+-----+---+-------------------------+---------------+----------------+ |1 |96 |107|heart murmur |Heart_Disease |Past | |1 |114 |127|echocardiogram |Test |Past | |2 |187 |200|echocardiogram |Test |Present | |2 |222 |227|normal |Test_Result |Present | |2 |229 |253|left ventricular function|Test |Present | |2 |260 |282|left atrial enlargement |Heart_Disease |Present | |3 |348 |361|ACE inhibitors |Drug_Ingredient|Planned | |3 |367 |375|diuretics |Drug_Ingredient|Planned |
Relation Extraction Result:
Relation extraction is a crucial task in NLP that involves identifying and classifying the semantic relations between entities mentioned in the text.
annotations = pipeline.fullAnnotate(text) rel_df = get_relations_df (annotations, 'relations') rel_df[rel_df.relation!="O"]
|sentence_id|relations |relations_entity1|relations_chunk1 |relations_entity2|relations_chunk2 | +-----------+-------------+-----------------+-------------------------+-----------------+-------------------------+ |1 |is_finding_of|Heart_Disease |heart murmur |Test |echocardiogram | |1 |is_date_of |Heart_Disease |heart murmur |Date |March 1998 | |1 |is_date_of |Test |echocardiogram |Date |March 1998 | |2 |is_finding_of|Test |echocardiogram |Heart_Disease |left atrial enlargement | |2 |is_result_of |Test_Result |normal |Test |left ventricular function| |2 |is_finding_of|Test |left ventricular function|Heart_Disease |left atrial enlargement |
In Spark NLP, .transform() is a method used to apply a trained NLP model to transform input data. This method takes the input data, such as text documents or data frames containing text, and applies the transformations defined by the NLP model. It is very efficient in large amounts of data.
NER Result:
data = spark.createDataFrame([["""
The patient admitted for gastrointestinal pathology, under working treatment.
History of prior heart murmur with echocardiogram findings as above on March 1998.
According to the latest echocardiogram, basically revealed normal left ventricular function with left atrial enlargement .
Based on the above findings, we will treat her medically with ACE inhibitors and diuretics and see how she fares.
"""]]).toDF("text")
result = pipeline.transform(data)
result.select(F.explode(F.arrays_zip(result.jsl_ner_chunk.result,
result.jsl_ner_chunk.begin,
result.jsl_ner_chunk.end,
result.jsl_ner_chunk.metadata)).alias("cols"))\
.select(F.expr("cols['0']").alias("chunk"),
F.expr("cols['1']").alias("begin"),
F.expr("cols['2']").alias("end"),
F.expr("cols['3']['entity']").alias("ner_label")).show()
+--------------------+-----+---+-------------------+ | chunk|begin|end| ner_label| +--------------------+-----+---+-------------------+ | admitted| 13| 20|Admission_Discharge| |gastrointestinal ...| 26| 51| Clinical_Dept| | heart murmur| 96|107| Heart_Disease| | echocardiogram| 114|127| Test| | March 1998| 150|159| Date| | echocardiogram| 186|199| Test| | normal| 221|226| Test_Result| |left ventricular ...| 228|252| Test| |left atrial enlar...| 259|281| Heart_Disease| | her| 328|330| Gender| | ACE inhibitors| 347|360| Drug_Ingredient| | diuretics| 366|374| Drug_Ingredient| | she| 388|390| Gender| +--------------------+-----+---+-------------------+
Assertion Status Result:
result.select(F.explode(F.arrays_zip(result.assertion_ner_chunk.result,
result.assertion_ner_chunk.begin,
result.assertion_ner_chunk.end,
result.assertion_ner_chunk.metadata,
result.assertion.result)).alias("cols"))\
.select(F.expr("cols['0']").alias("chunk"),
F.expr("cols['1']").alias("begin"),
F.expr("cols['2']").alias("end"),
F.expr("cols['3']['entity']").alias("ner_label"),
F.expr("cols['4']").alias("assertion")).show()
+--------------------+-----+---+---------------+---------+ | chunk|begin|end| ner_label|assertion| +--------------------+-----+---+---------------+---------+ | heart murmur| 96|107| Heart_Disease| Past| | echocardiogram| 114|127| Test| Past| | echocardiogram| 186|199| Test| Present| | normal| 221|226| Test_Result| Present| |left ventricular ...| 228|252| Test| Present| |left atrial enlar...| 259|281| Heart_Disease| Present| | ACE inhibitors| 347|360|Drug_Ingredient| Planned| | diuretics| 366|374|Drug_Ingredient| Planned| +--------------------+-----+---+---------------+---------+
For more pre-trained pipelines, you can visit the Spark NLP Models Hub page.
Using pre-trained pipelines offers several advantages over building a custom NLP pipeline:
- Time Efficiency: Pretrained pipelines are readily available and require minimal setup, saving time compared to developing a custom pipeline from scratch.
- Quality Assurance: Pretrained pipelines are often developed and maintained by experts, ensuring high-quality performance and reliability in various NLP tasks.
- State-of-the-Art Performance: Pretrained pipelines incorporate cutting-edge techniques and models, providing access to state-of-the-art performance without the need for extensive experimentation and research.
- Scalability: Pretrained pipelines are designed to handle large-scale data processing efficiently, making them suitable for deployment in production environments with high-volume data streams.
- Cost-Effectiveness: Using pre-trained pipelines can be more cost-effective than investing resources in building and maintaining a custom NLP pipeline, especially for organizations with limited budgets or expertise in NLP.
Overall, leveraging pre-trained pipelines offers a convenient and effective way to incorporate advanced NLP capabilities into applications and workflows, with benefits including time savings, high performance, and scalability.
Conclusion
In conclusion, pre-trained pipelines greatly simplify the process of understanding and interpreting clinical or medical records. Instead of creating a long, complicated pipeline, we can accomplish the same task with just one-liner code. This saves time and effort when dealing with complex documents such as healthcare forms. The speed, efficiency, and operational benefits provided by pre-trained pipelines will play an important role in data analysis efforts in the healthcare industry.