What is De-Identification in Medical Images?
Healthcare organizations generate and manage enormous amounts of sensitive patient information from hospital records and clinical notes to high-resolution medical images that capture intimate details of a person’s health.
As care becomes increasingly digital and interconnected, this information is routinely shared between hospitals, specialists, diagnostic centers, research institutions, and even patients themselves. Medical images, stored and exchanged in the DICOM format, play a central role in diagnosis and treatment; however, they also introduce unique privacy challenges: they are large, complex, require specialized viewers, and often contain both metadata-level and pixel-level identifiers that cannot be shared without proper protection.
In an era where compliance frameworks like HIPAA, GDPR, CCPA, and CPRA hold organizations accountable for protecting personal data, robust de-identification has become essential. It allows healthcare providers and researchers to unlock the full value of medical imaging, enabling AI development, clinical research, multi-site collaboration, and large-scale data analysis while ensuring patient privacy remains uncompromised.
Challenges with De-Identification of Medical Images
The task of DICOM de-identification is overwhelmingly complex because it demands a comprehensive scrubbing across thousands of standard and private tags with countless Value Representation (VR) permutations, while simultaneously locating and erasing PHI burned into the pixel data as overlays or annotations. This must be executed consistently across multi-frame objects and entire clinical studies, necessitating the generation of new, yet coherent, Study and Series UIDs. Without a dedicated, automated solution, this manual process is virtually guaranteed to introduce errors, compromise patient privacy through missed PHI, and violate regulatory compliance, making the use of a specialized de-identification tool essential for safe and effective data sharing.
John Snow Labs Visual NLP De-Identification Solution for Medical Images
In this blog post, we’ll walk through a full step-by-step pipeline build using John Snow Labs’ Visual-NLP models to tackle these challenges: reading images & metadata tags, detecting sensitive content in pixels and metadata, transforming or removing it, and validating your output. To make things more interesting, we’ll use the recently released MIDI-B Dataset.
Beyond simple removal, the pipeline also supports full obfuscation of PHI in both the metadata and the pixel levels of DICOM files. Metadata fields can be masked, shifted, hashed, or replaced with consistent synthetic values, ensuring that identifiers remain unusable while keeping the structural integrity needed for downstream analysis. Pixel regions containing text or overlays such as names, dates, IDs, or accession numbers can be detected and selectively blurred, masked, or replaced, allowing the visual context of the image to remain intact while eliminating sensitive content. This dual-layer obfuscation ensures that datasets remain clinically meaningful while fully protecting patient privacy.
Introduction To Visual NLP
John Snow Labs Visual-NLP lies at the intersection of Computer Vision and Natural Language Processing, tailored for medical imaging; it includes models that can understand image content and associated text (both visible pixel annotations and metadata tags).

Features of Visual NLP
In the DICOM context, Visual-NLP lets you:
- Detect text regions in image pixels.
- Extract text from Pixel PHI regions.
- Recognize and classify that text (patient name, Phone Number, Address, ID, Date)
- Generate Coordinates for sensitive text regions for Pixel-level masking.
- Read DICOM metadata tags and categorize them (public and private)
- Make informed decisions on whether to keep, remove, obfuscate or replace any sensitive content present in the metadata sources.
- Render a new DICOM file with original characteristics, but free from all Patient PHI.
All Visual-NLP DICOM de-identification flows consist of two main parts: Ingestion and Finalizer.
Pixel De-Identification
For Pixel De-Identification, we extract the image objects directly from the DICOM file and run text detection, text recognition, NER detection, and coordinate generation to locate every PHI region. In Flow 1, DicomToImageV3 serves as the ingestion stage, while DicomDrawRegions acts as the finalizer. The output of this process is a fully reconstructed DICOM file where all PHI has been removed at the pixel level.
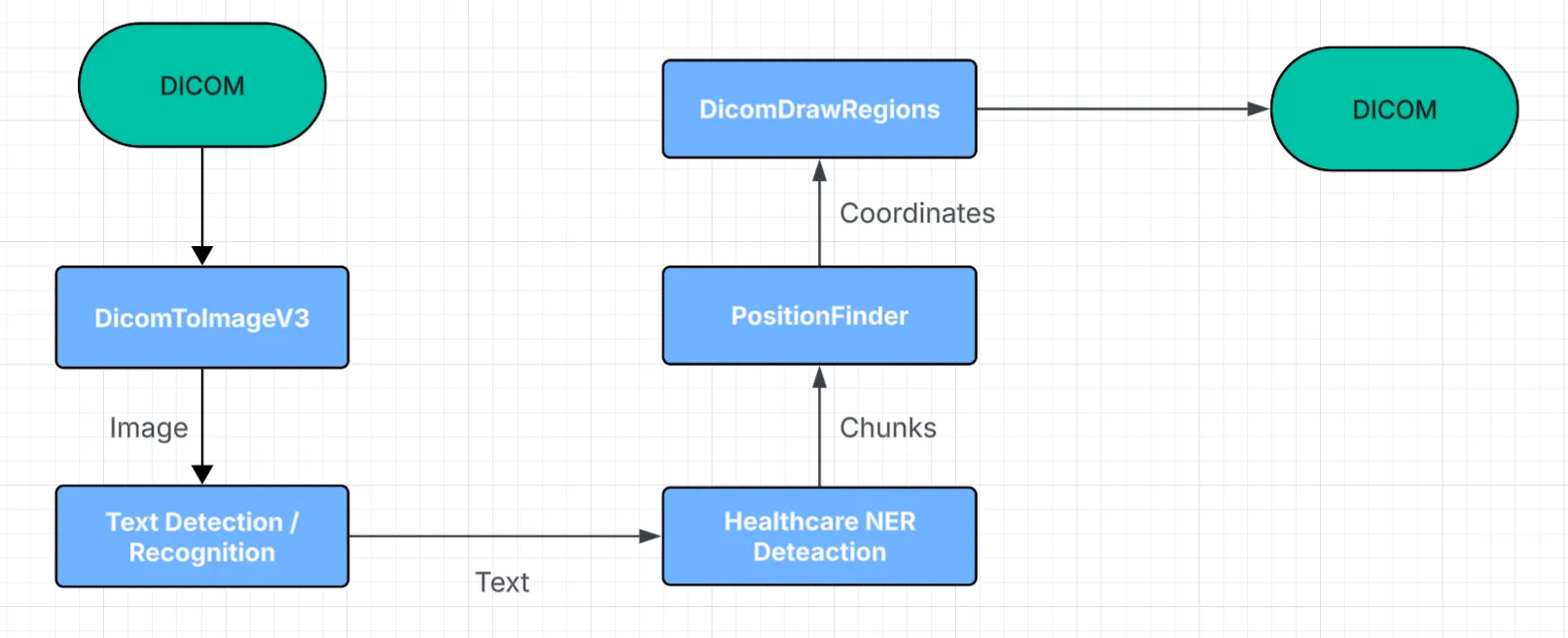
Flow 1 Pixel De-Identification
dicom_to_image = DicomToImageV3() \
.setInputCols(["content"]) \
.setOutputCol("image_raw") \
.setFrameLimit(0) \
.setKeepInput(False) \
.setCompressionThreshold(1) \
.setCompressionQuality(95) \
.setCompressionMode("auto") \
.setMemoryOptimized(True)
Input: DICOM Path/ Spark Binary Object
Output: Visual NLP Image Object
Parameters:
- setFrameLimit ( Type: Integer, Default: 0 ) -> Limit number of frames for extraction. To extract all frames, set this parameter to 0.
- setScale ( Type: Float, Default: 0 ) -> Width of the desired input image. Image will be resized to this width.
- setCompressionThreshold ( Type: Integer, Default: 1 ) -> Number of Mega Pixels in an image required to trigger compression if compressionMode==auto.
- setMemoryOptimized ( Type: Boolean, Default: False ) -> Whether to perform memory optimised transform.
- setCompressionQuality ( Type: Integer, Default: 85 ) -> JPEG quality of compression if applied, value between 1 and 95
- setCompressionMode ( Type: String, Default: auto ) -> Perform compression of the DICOM file internally, possible values auto/enabled/disabled.
- setKeepInput ( Type: Boolean, Default: True ) -> Flag indicating whether the input column should be retained in the DataFrame.
DicomDrawRegions (Finalizer)
This stage takes a DICOM path or object along with a set of coordinates and accordingly masks the specified regions in the image. Beyond masking, it also performs full reconstruction of the DICOM file, ensuring the output is rendered with the correct transfer syntax, appropriate pixel compression, and all image-related DICOM tags updated to reflect the new compression and image characteristics.
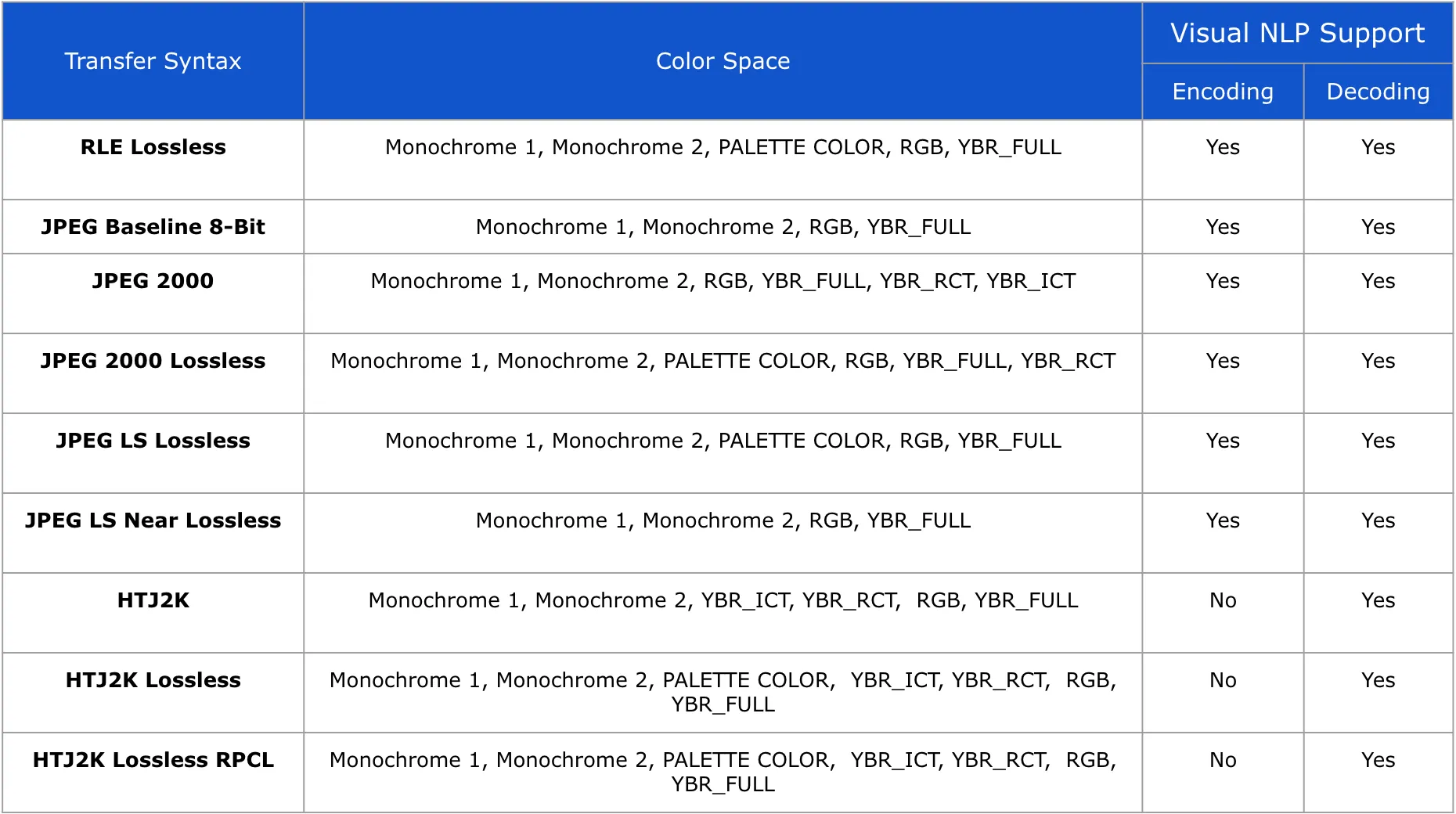
Support for Transfer Syntaxes in DicomDrawRegions
Visual NLP does not alter the Photometric Interpretation / Color Space of DICOM pixels in any way. The final DICOM file retains the same characteristics as the input.
draw_regions = DicomDrawRegions() \
.setInputCol("dicom_metadata_cleaned") \
.setInputRegionsCol("coordinates") \
.setOutputCol("dicom_final") \
.setAggCols(["path"]) \
.setKeepInput(False)
Input: DICOM Path/ Spark Binary Object, Coordinates
Output: DICOM Object
Note : This is an aggregation stage, it groups data by path collecting coordinates from all the frames to DeIdentify the Pixels and generate the final DICOM File. All intermediate results prior to this stage will be lost.
Parameters:
- setScaleFactor ( Type: Float, Default: 1) -> If the Image Object extracted from DicomToImageV3 was scaled up/down, this means that all the coordinates that are generated should also be scaled to fit the original image dimensions.
- setInputRegionsCol ( Type: String, Default: regions ) -> Dataframe column containing Coordinates generated for the PHI present in the pixels, generated from PositionFinder.
- setMemoryOptimized ( Type: Boolean, Default: False ) -> Flag indicating whether to perform memory optimised transform.
- setForceOutput ( Type: Boolean, Default: False ) -> Flag to indicate whether to force output for DICOM files for which input compression is not supported.
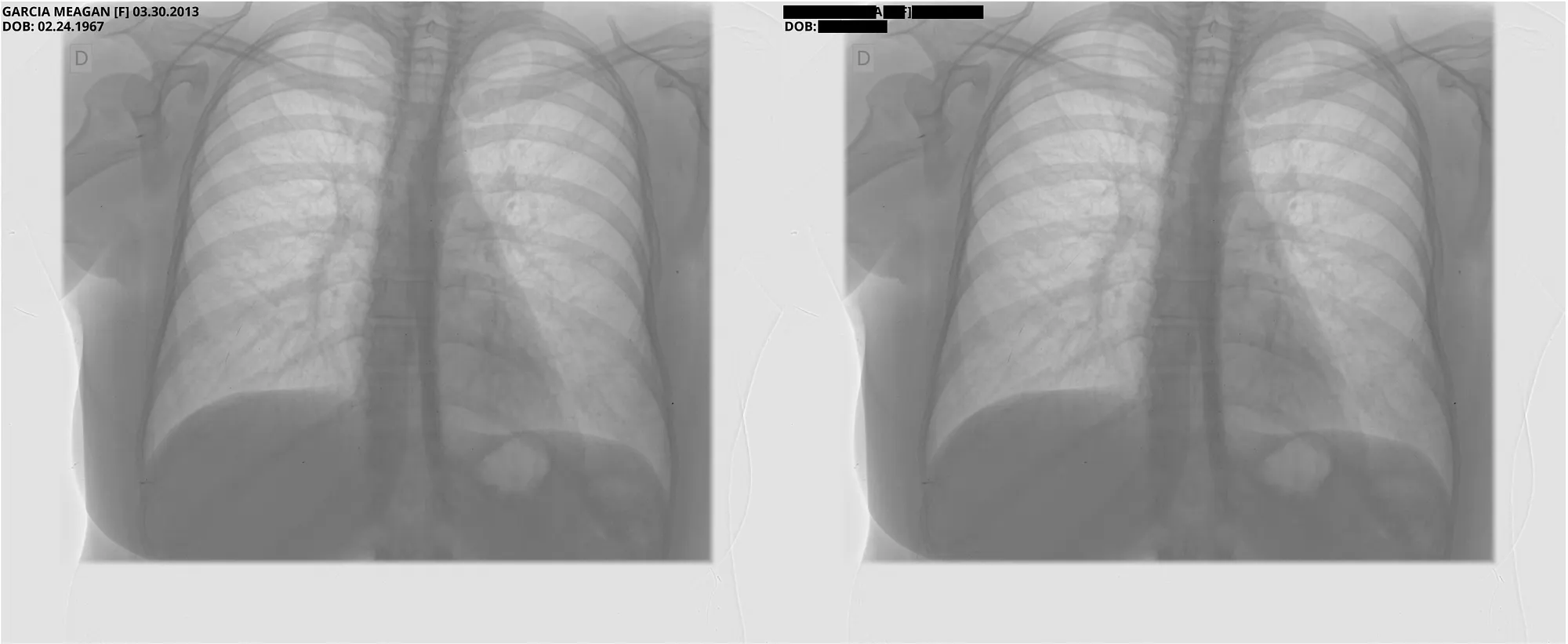
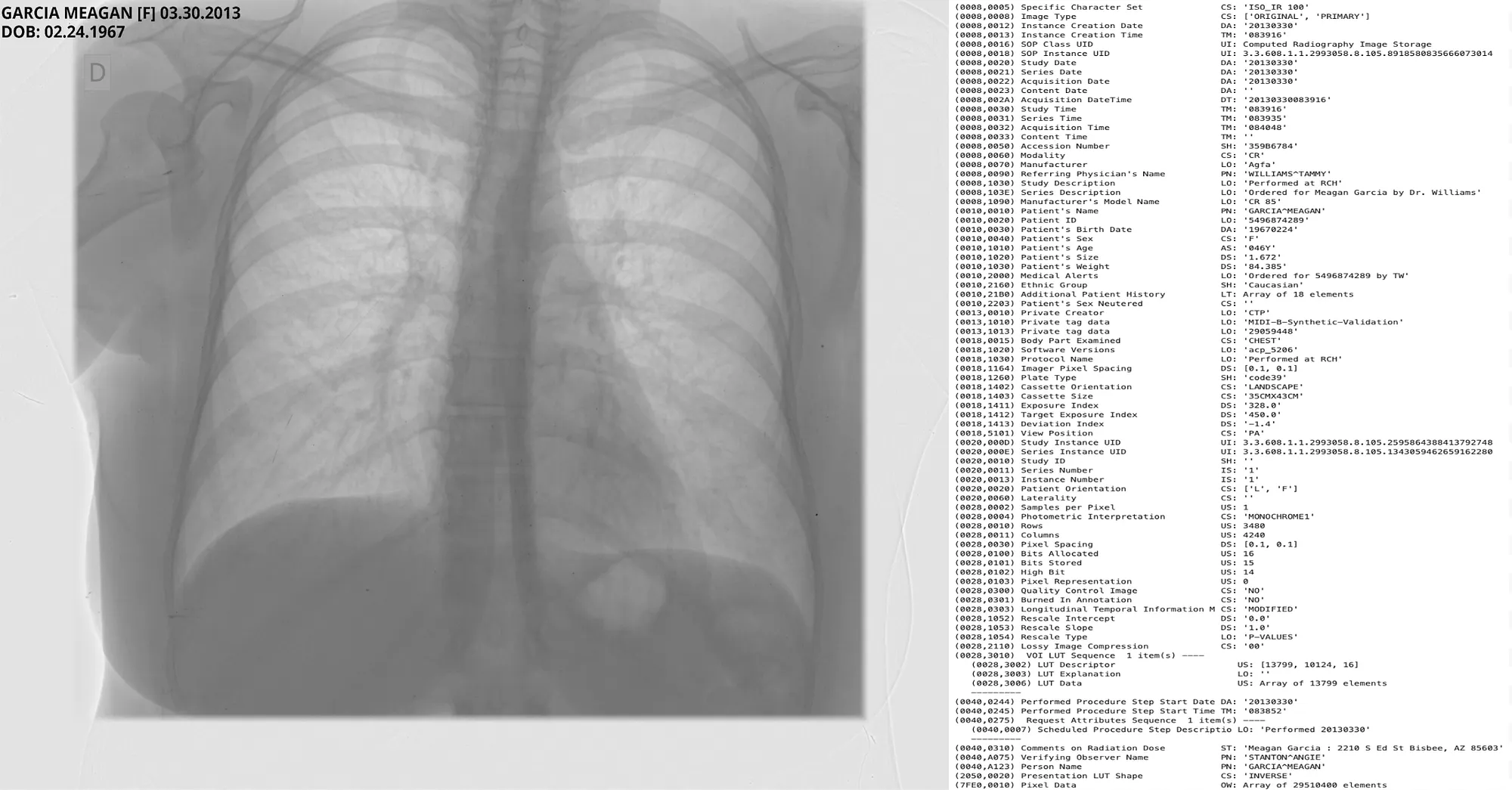
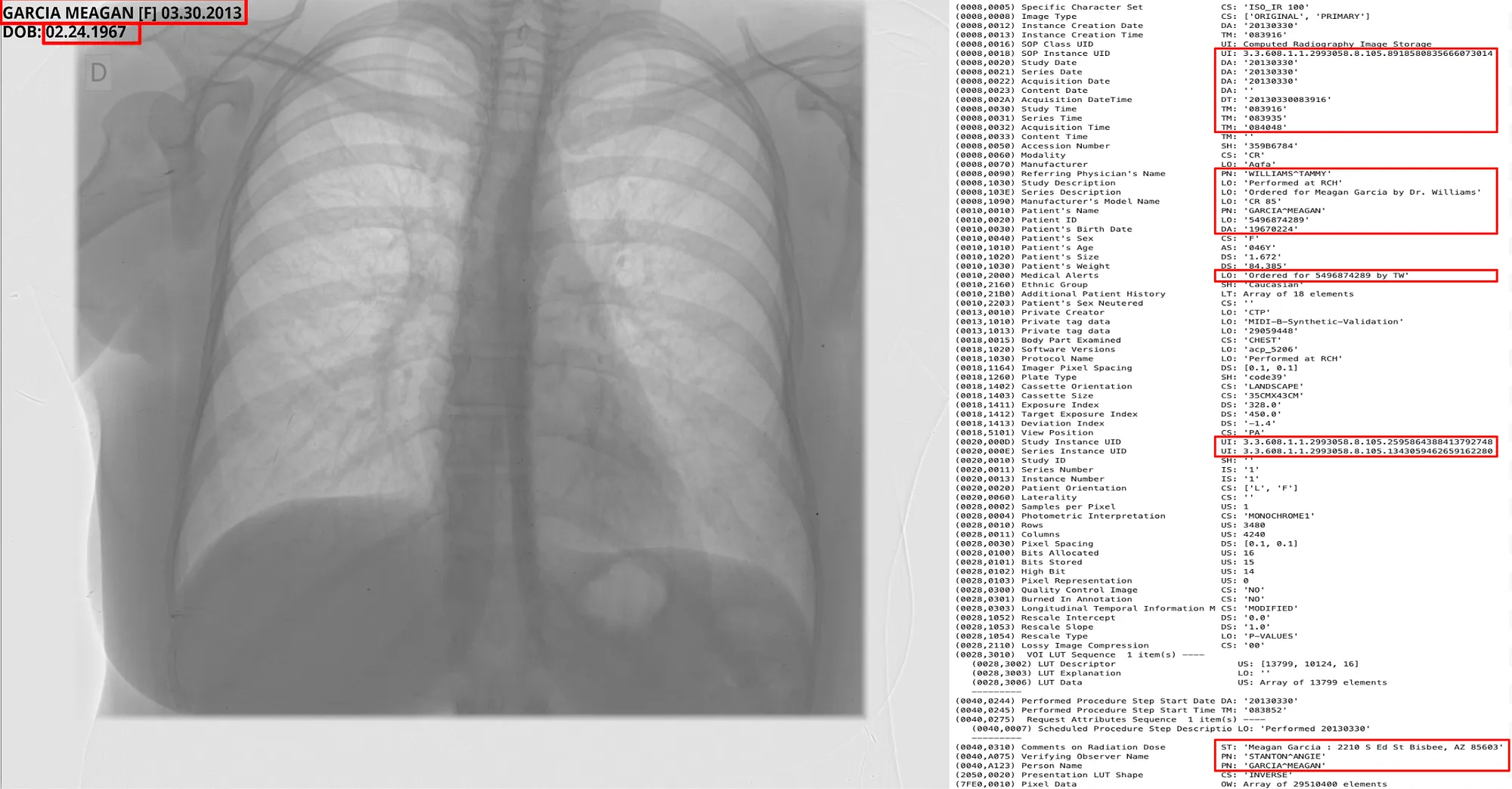
Pixel DeIdentification Original ( Left ) -> DeIdentified ( Right )
Reference : MIDI-B Pixel Metadata DeIdentification Notebook
Metadata/Tag De-Identification
For Metadata De-Identification, we first extract the Metadata, the Document Object, and the tag-mapping only for those tags marked with the cleanTag action in the Strategy file. This is what happens in Flow 2. For every other action in the Strategy file, the metadata alone provides everything we need, so no extra steps are required; these fall under Flow 3. In this setup, the ingestion stage is DicomToMetadata, and the finalisation stage is DicomMetadataDeIdentifier.
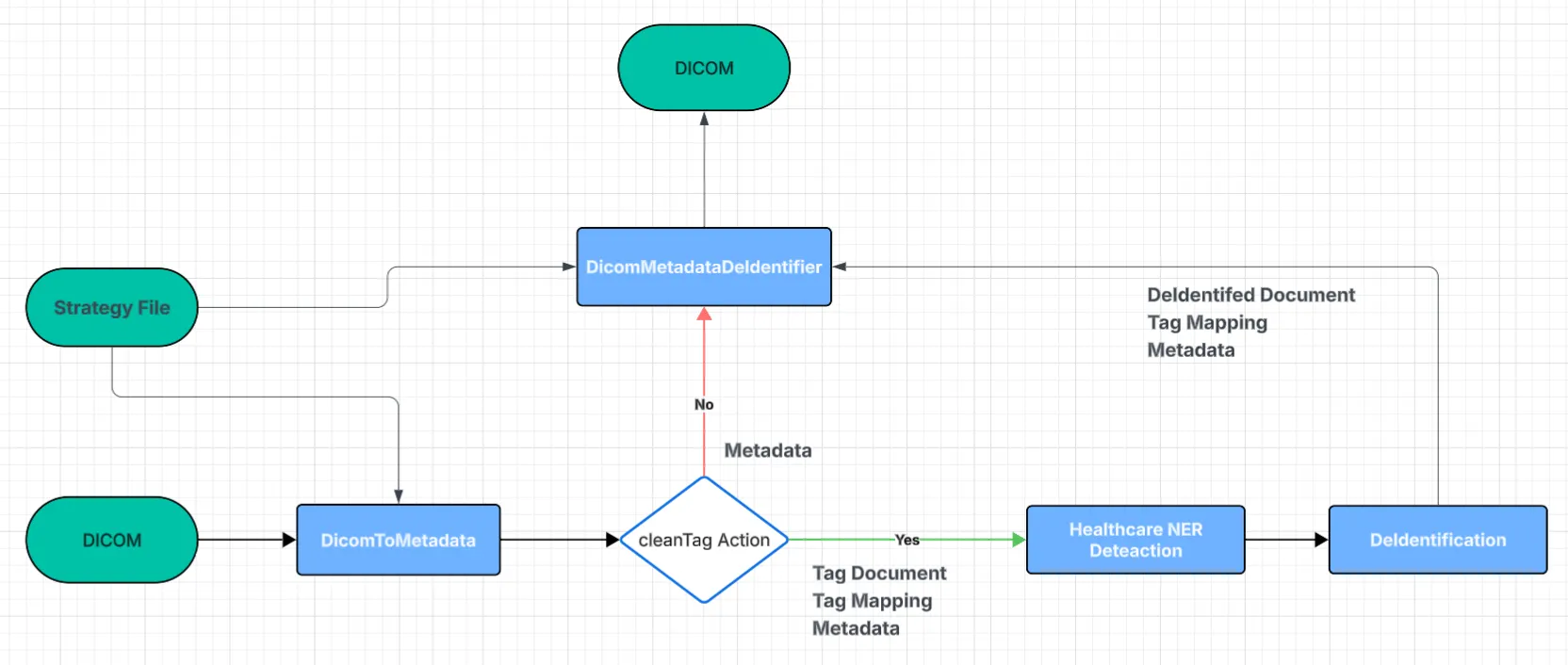
Flow 2 & 3 Metadata De-Identification
DicomToMetadata (Ingestion)
Extract the full metadata from the DICOM file as a JSON string for each tag present. Using the strategy file, identify all tags marked with the cleanTag action and retrieve their corresponding Document Object and tag-mapping information. This stage also supports reading DICOM files directly from cloud storage services such as Amazon S3.
dicom_to_metadata = DicomToMetadata() \
.setInputCol("path") \
.setOutputCol("metadata") \
.setKeepInput(True) \
.setExtractTagForNer(True) \
.setTagMappingCol("tag_mapping") \
.setDocumentCol("t_document") \
.setStrategyFile(strategy_file_path)
Input: DICOM Path / Spark Binary Object
Output: Metadata, Document Object ( Optional ), Tag Mapping ( Optional )
Every metadata tag assigned the cleanTag action is transformed into a Document object. These Documents act as the core tag-level inputs for all downstream Healthcare and Visual NLP components.
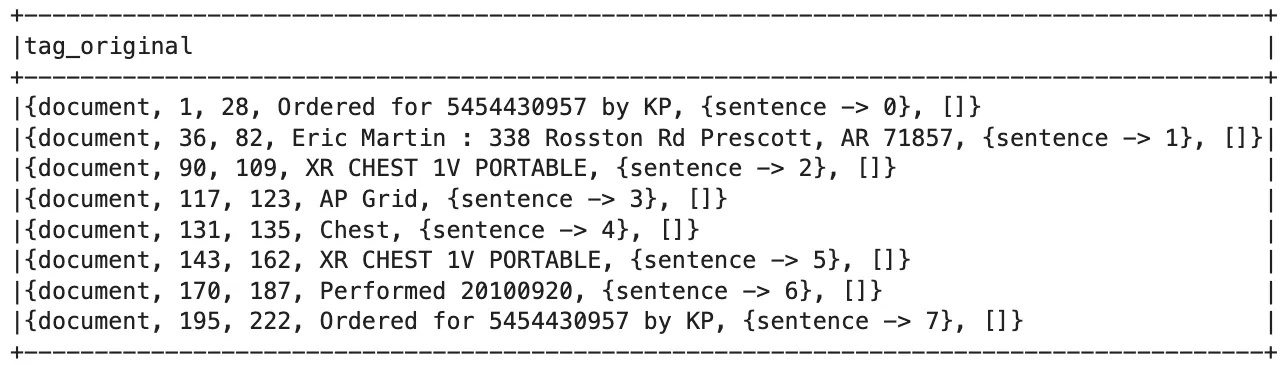
DICOM Document Object
Parameters:
- setDocumentCol ( Type: String, Default: dicom_document ) -> DataFrame column name which will contain the output DICOM Document Object.
- setTagMappingCol ( Type: String, Default: tag_mapping ) -> DataFrame column name which will contain the output DICOM Tag Mapping for the DICOM Document Object.
- setExtractTagForNer ( Type: Boolean, Default: True ) -> Flag indicating whether DICOM Document Object and Tag Mapping should be extracted. If False, will only extract the metadata. Set to True to remove PHI using NER.
- setKeepInput ( Type: Boolean, Default: True ) -> Flag indicating whether the input column should be retained in the DataFrame.
- setStrategyFile ( Type: String ) -> Path to strategy file; all tags associated with the cleanTag action will be extracted for NER.
- setOutputCol ( Type: String, Default: metadata ) -> DataFrame column name that will contain the DICOM metadata as a JSON String.
DicomMetadataDeIdentifier (Finalizer)
Metadata is always passed as the default input, while the de-identified document-tag object and tag-mapping are optional inputs. Using these, the component removes PHI from all tags marked with the cleanTag action, and for all other actions, it applies the appropriate transformation required to de-identify the tag’s value. The result is a new DICOM file with fully cleaned and updated metadata.
dicom_deidentifier = DicomMetadataDeidentifier() \
.setInputCols(["path", "metadata"]) \
.setOutputCol("dicom_metadata_cleaned") \
.setTagMappingCol("tag_mapping") \
.setKeepInput(True) \
.setTagCleanedCol("deid_documents") \
.setStrategyFile(strategy_file_path)
Input: DICOM Path/ Spark Binary Object, Tag Mapping, DeIdentified Document Object
Output: DICOM Object
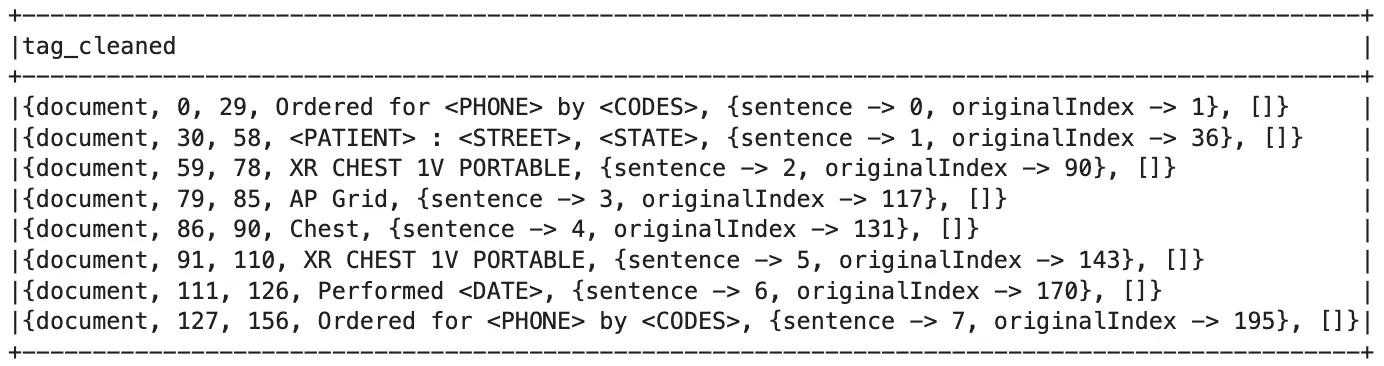
De-Identified Document Object
Parameters:
- setTagMappingCol ( Type: String, Default: tag_mapping ) -> DataFrame col containing tag mapping values generated from DicomToMetadata stage.
- setTagCleanedCol ( Type: String, Default: tag_mapping ) -> DataFrame col containing Document Object with PHI removed, generated from DeIdentification stage.
- setRegenerateSeries ( Type: Boolean, Default: False ) -> Flag to indicate whether Series Number should be regenerated deterministically.
- setRemovePrivateTags ( Type: Boolean, Default: False ) -> Flag indicating whether private tags should be removed from metadata.
- setStrategyFile ( Type: String ) -> Path to strategy file, all tags associated with the cleanTag action will be extracted for NER.
- setKeepInput ( Type: Boolean, Default: True ) -> Flag indicating whether the input column should be retained in the DataFrame.
The Strategy file is the core of the entire metadata de-identification system. It decides which tags to clean, which ones to keep, and how each part of the data should be processed.
After working with the MIDI-B dataset, we created two versions of this file: one with all the tags we discovered and cleaned during our experiments, and another that includes everything from MIDI-B plus additional tags needed for TCIA.
Tags→ DICOM tag ID in hexadecimal (group, element).VR→ Data type of the tag.Name→ Human-readable tag name.Status→ Indicates if the tag is retired; use “Retired” if applicable.Action→ De-identification operation to perform.Option→ Optional value for actions like replaceWithLiteral or shiftDateByFixedNbOfDays.
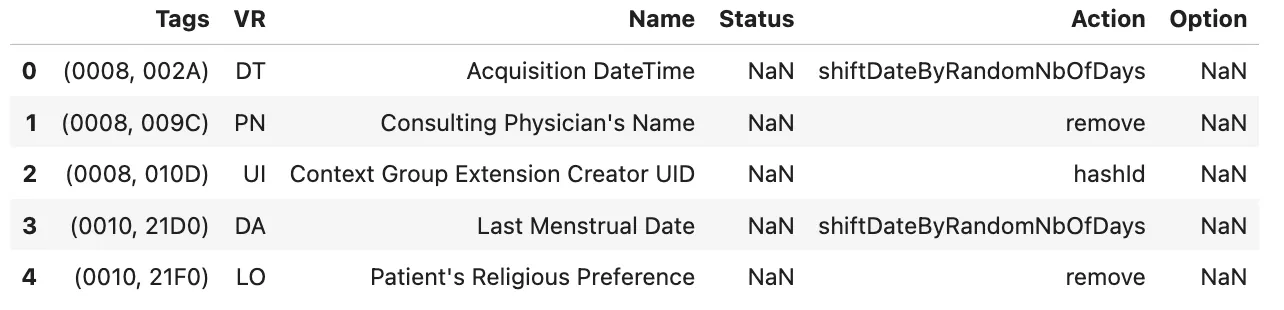
Example Rows from Strategy File
As an end user, you can define your own tags and choose exactly what you want to do with them using the available actions. Each action applies only to specific VR types, and together the tag and its VR form the primary key that determines how that piece of metadata will be handled. Below, you can find all the supported actions (allowed VR) and their descriptions.
replaceWithRandomName ( PN, LO )→ Replaces person names with randomly generated obfuscated names.replaceWithLiteral ( ALL )→ Substitutes a tag’s value with a fixed literal string (e.g., “REDACTED” or “REMOVED”).cleanTag ( ALL )→ Collects DICOM tags and converts them into a document-like format, an entry point for Healthcare NLP stages, preparing tag values for PHI detection using NER models or Regex rules.delete ( ALL )→ Deletes the DICOM tag entirely from the file.remove ( ALL )→ Clears the tag’s value but leaves the tag key in place.ensureTagExists ( ALL )→ Creates the tag if it’s missing with defaults.patientHashId ( LO )→ Hashes the original Patient ID to create a new consistent anonymised ID of the same length.hashId ( UI, LO, SH )→ Hashes the original UID to create a new, consistent anonymised UID.shiftUnixTimeStampRandom ( SL, FD )→ Randomly shifts Unix timestamps.shiftDateByRandomNbOfDays ( DA, DT )→ Offsets dates by a random number of days.shiftDateByFixedNbOfDays ( DA, DT )→ Moves all dates by a fixed number of days for deterministic values.shiftTimeByRandom ( TM )→ Randomly shifts recorded time values.shiftAgeByRandom ( AS )→ Randomly adjusts recorded age values.
To help you visualise, we will use a sample DICOM file from the MIDIB dataset, which contains PHI in the metadata and Pixels.

Metadata DeIdentification Original ( Left ) -> DeIdentified ( Right )
MIDI-B Dataset Overview
The MIDI-B challenge follows three major frameworks that together define what a compliant, privacy-safe DICOM dataset looks like:
- HIPAA Safe Harbor (U.S.)
Under the HIPAA Privacy Rule, Protected Health Information (PHI) must be stripped of any details that could identify a patient. The “Safe Harbor” method lists 18 types of identifiers ( from names, dates, and contact info to IP addresses, biometric data, and full-face images) that must be removed or obscured. Once these are gone, the data can be safely shared for research without revealing anyone’s identity. - DICOM PS3.15 (Attribute Confidentiality Profile)
The DICOM standard itself provides detailed rules for protecting sensitive metadata inside medical images. PS3.15 specifies which tags should be deleted, replaced, or anonymized, and how to handle UIDs so that related studies remain linkable for research. It ensures that both metadata and image pixels remain compliant when data is transmitted or shared. - TCIA Submission Guidelines
The Cancer Imaging Archive (TCIA) adds another layer of best practices for preparing datasets for public release. These guidelines focus on verifying that all PHI is removed from both metadata and from the image itself and that the dataset maintains consistency and research value after de-identification.
Together, these standards form the backbone of modern medical image de-identification. They ensure patient privacy is preserved while keeping the data scientifically useful for machine learning, diagnostics, and benchmarking efforts like MIDI-B.
Due to its size, variety, and complexity (subjects, modalities, studies), the MIDI-B dataset is both a realistic production-scale challenge and an ideal proving ground for advanced de-identification pipelines.
The dataset is organized into Validation and Test sets, each containing two subsets: a Synthetic set, where PHI has been artificially inserted into tags and pixel regions, and a Curated set, where the same files have been manually de-identified to serve as the ground truth.
For de-identification, MIDI-B checks for the following actions
tag_retained→ Verifies that required non-PHI DICOM tags were correctly preserved after de-identification.text_notnull→ Checks that text fields expected to contain information are not empty in the output.text_retained→ Confirms that non-sensitive text content was successfully kept.text_removed→ Ensures that sensitive or PHI text detected in pixels or metadata was properly removed.date_shifted→ Validates that original date values were offset or randomized while maintaining relative time gaps.uid_changed→ Confirms that all UIDs were replaced with new, consistent identifiers.pixels_retained(Pixels) → Checks that images without PHI were preserved without alteration.uid_consistent→ Ensures that newly generated UIDs remain consistent across related DICOM objects.patid_consistent→ Confirms that Patient IDs are anonymized but still linked correctly within the same patient’s studies.pixels_hidden(Pixels) → Measures whether PHI-containing regions in image pixels were fully hidden or obfuscated.
MIDI-B Validation Results
The MIDI-B Validation Script produces two complementary outputs during the DICOM de-identification evaluation process. The first is a validation database, which contains detailed records of all validation checks performed on each DICOM file. The second output is an Excel-based Pixel Masking Report that provides an overview of all PHI masked within the pixel data.
PHI Formats in Pixel Data:
- Typical structure: FirstName LastName [Gender] Date\nDate
- Codes considered as PHI: JT, SWU, JKR, MWF, ICG, NKF, YH
Validation Results:
Validation Pixel Masking Excel Report: Link
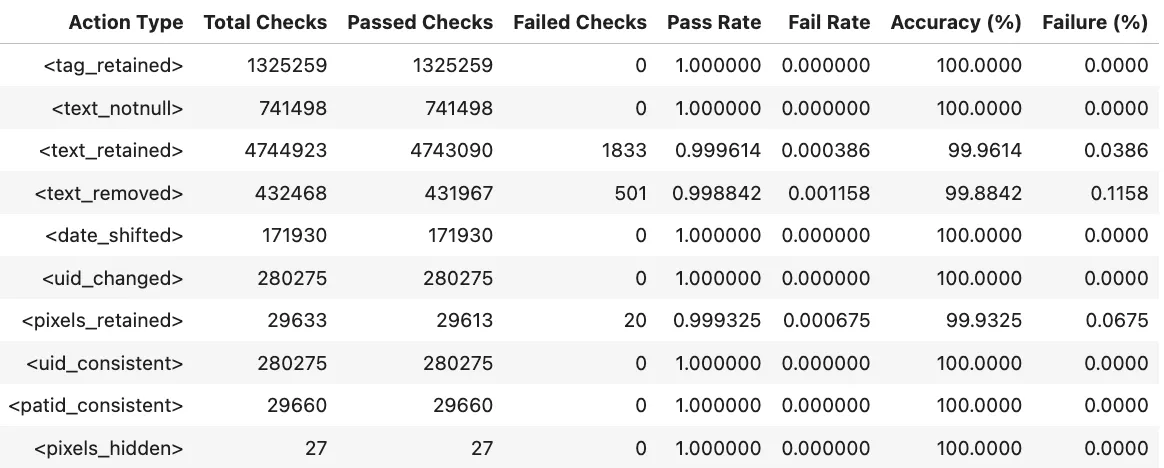
Validation Report Summary (Validation Set)
Test Results:
Test Pixel Masking Excel Report: Link
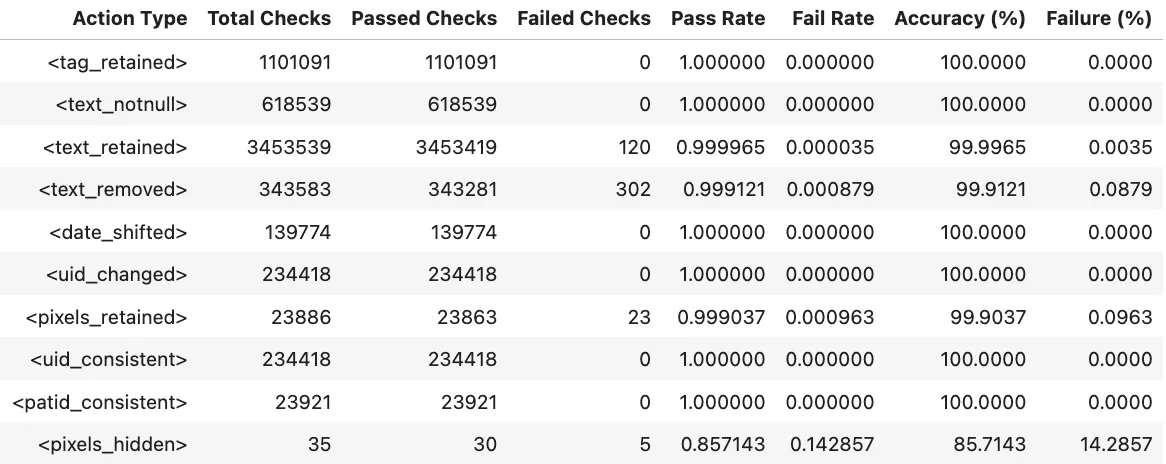
Validation Report Summary (Test Set)
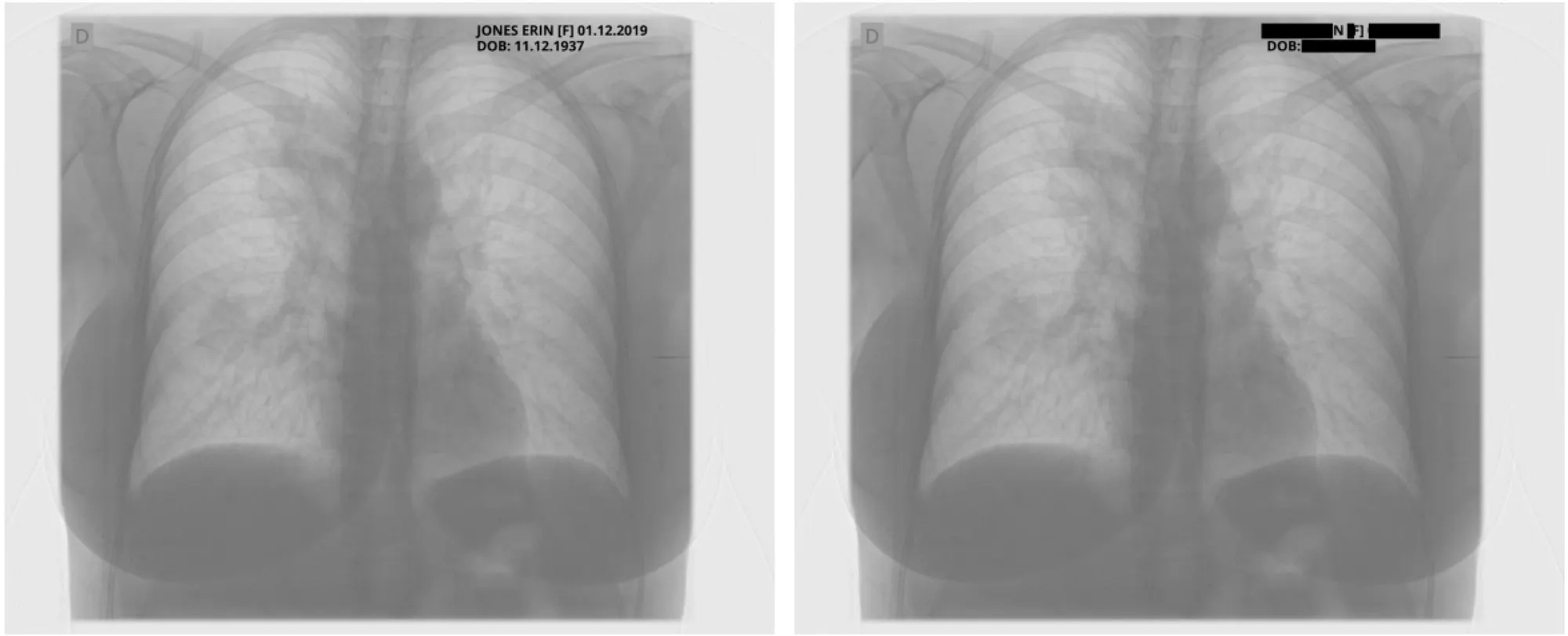
Sample Pixel Result 1
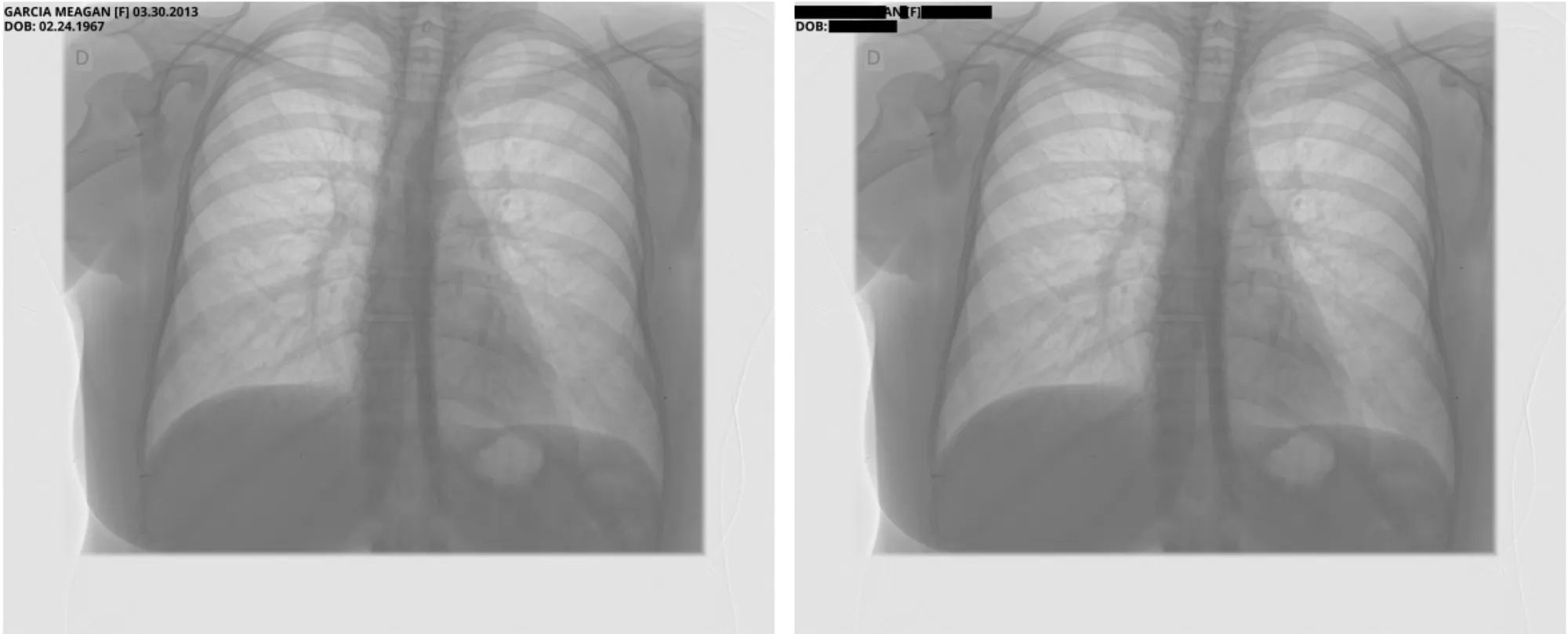
Sample Pixel Result 2
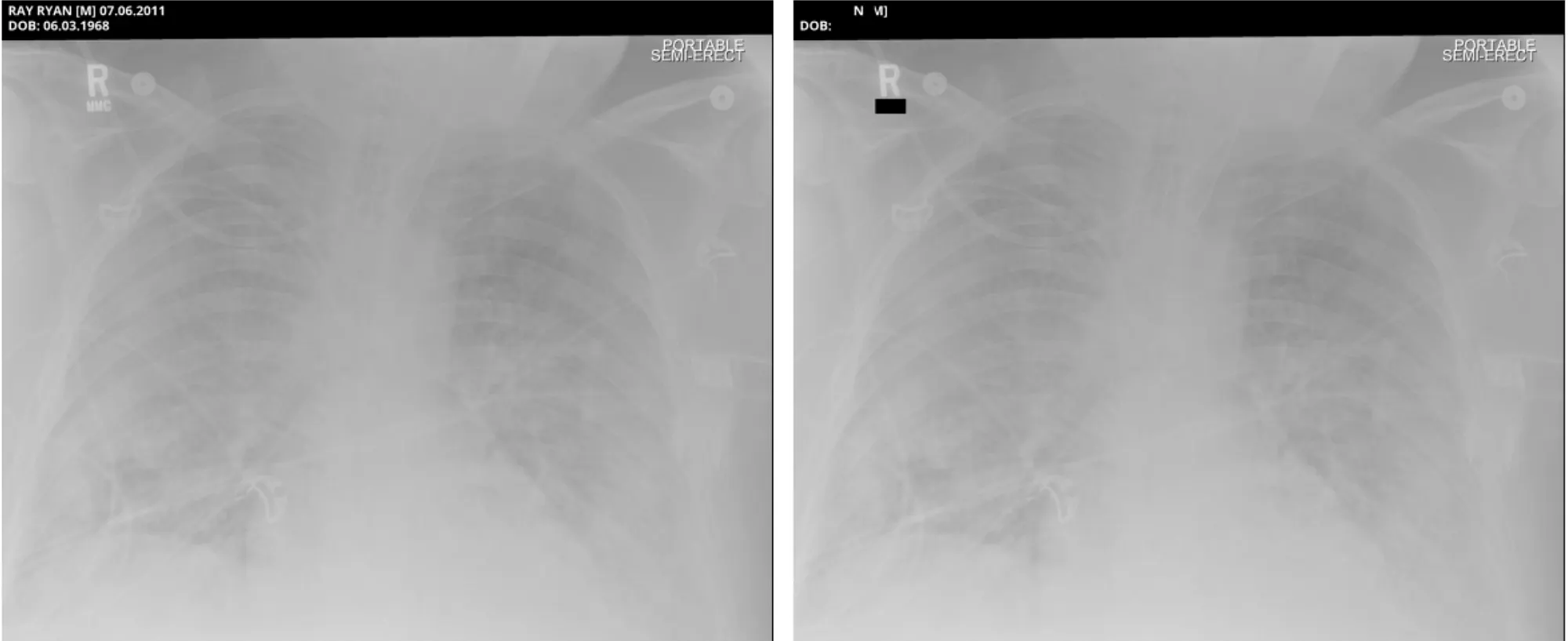
Sample Pixel Result 3
Reference: A detailed walkthrough of the Pixel De-Identification process, including how OCR can be used for before and after de-identification to evaluate Pixel PHI masking within the MIDI-B Subset.
Conclusion
The MIDI-B dataset has been invaluable in measuring and validating the quality of DICOM de-identification.
With John Snow Labs Visual NLP, organizations can finally de-identify DICOM images end-to-end using a single, unified, and fully configurable pipeline. By combining pixel masking, metadata cleaning, PHI-aware NER models, and complete DICOM Metadata/Pixel reconstruction, the solution delivers enterprise-grade protection.
The strategy-file architecture makes the system exceptionally adaptable: every tag action, whether hashing, removing, replacing, updating, or cleaning, is controlled through a simple configuration rather than code changes. This gives users precise, consistent, and maintainable de-identification behavior across slices.
In short, John Snow Labs Visual NLP enables healthcare providers, data platforms, and AI teams to share and operationalize imaging data safely and confidently, with a product designed for real-world scale, strict compliance requirements, and modern medical AI workflows.
References