As healthcare organizations increasingly adopt Large Language Models (LLMs) and Vision Language Models (VLMs) for clinical applications, the question of “how to deploy these models securely and efficiently” becomes critical. At John Snow Labs, we’ve worked closely with Databricks to provide three flexible deployment options that balance performance, control, and ease of use — all while maintaining the security and compliance standards that healthcare demands.
In this post, I’ll walk you through each deployment option based on a recent customer demonstration, showing you exactly how to leverage our state-of-the-art medical LLMs on Databricks infrastructure.
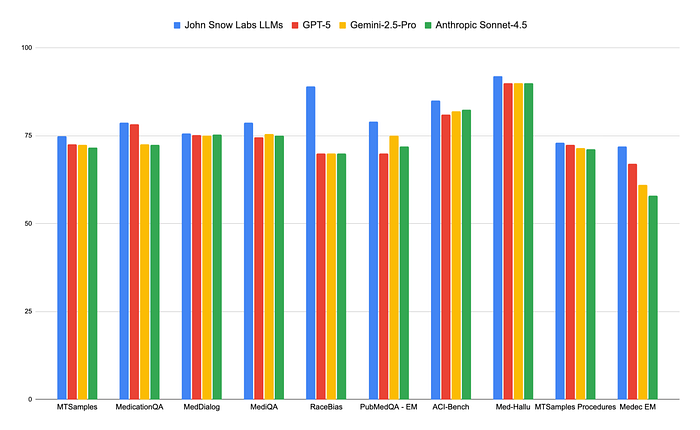
Why Medical-Specific LLMs Dramatically Outperform General Models
Before diving into deployment options, let’s establish why specialized medical LLMs aren’t just incrementally better — they’re transformationally superior for healthcare applications.
OpenMed Benchmark Results
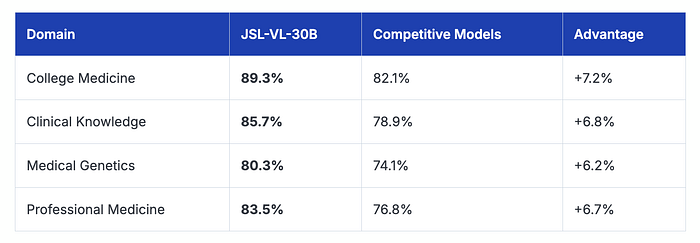
Our JSL-VL-30B (Vision Language Model) achieves 83.5% average accuracy across critical medical domains
Multi-Modal Medical Benchmark: Vision + Reasoning
For vision-based medical understanding tasks
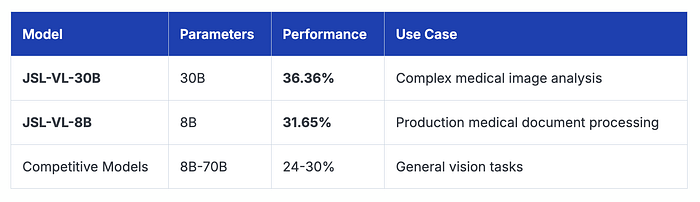
Our models show 15–50% better performance on tasks requiring visual medical reasoning — critical for processing medical images, charts, and handwritten notes.
MedHELM: Healthcare-Specific Evaluation
On Stanford’s MedHELM benchmark (healthcare-specific evaluation):
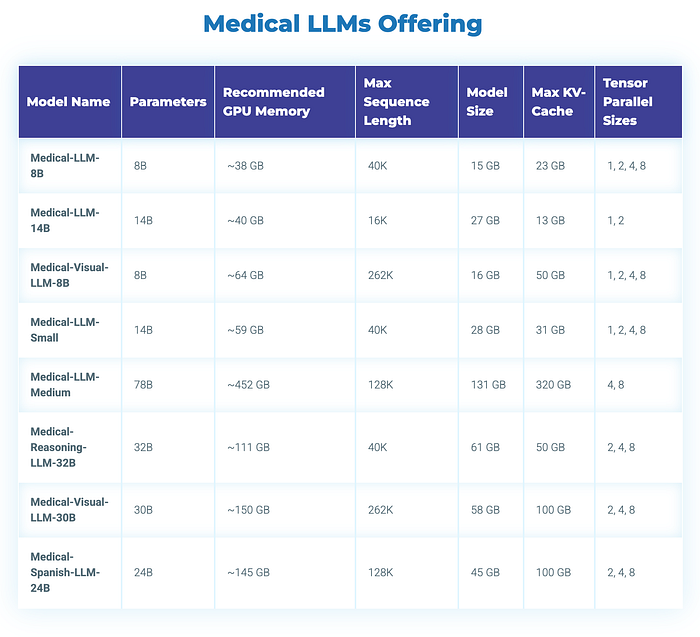
John Snow Labs Medical LLMs Offerings at Marketplaces
There is overwhelming evidence from both academic research and industry benchmarks that domain-specific, task-optimized large language models consistently outperform general-purpose LLMs in healthcare. At John Snow Labs, we’ve developed a suite of Medical LLMs purpose-built for clinical, biomedical, and life sciences applications.
Our models are designed to deliver best-in-class performance across a wide range of medical tasks — from clinical reasoning and diagnostics to medical research comprehension and genetic analysis.
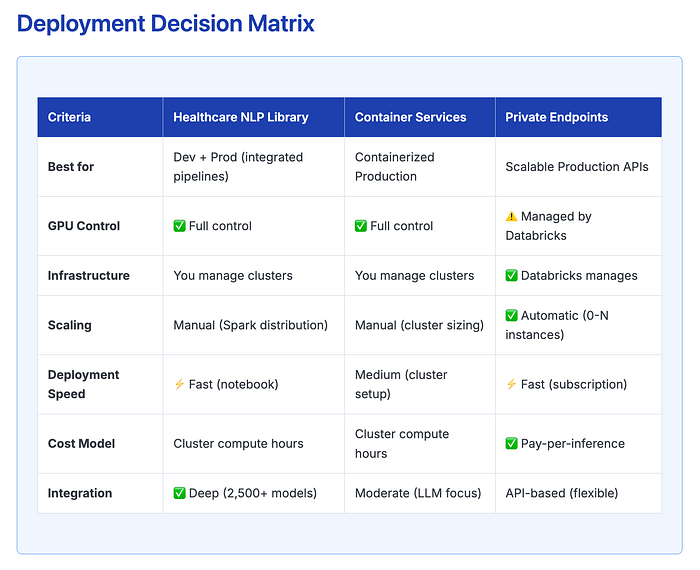
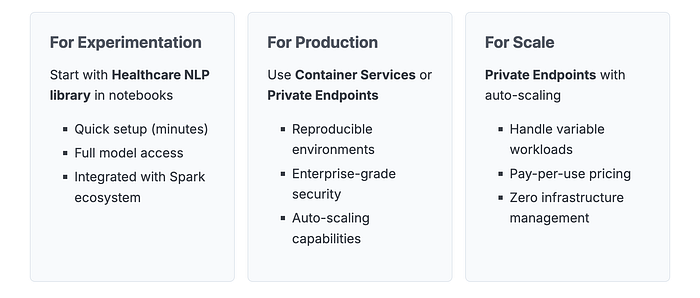
Three Deployment Options on Databricks
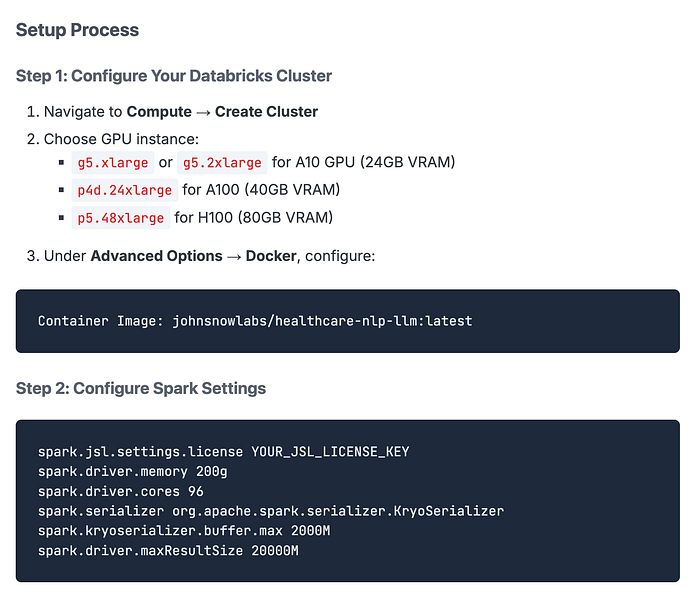
Option 1: Healthcare NLP Library (Direct Integration)
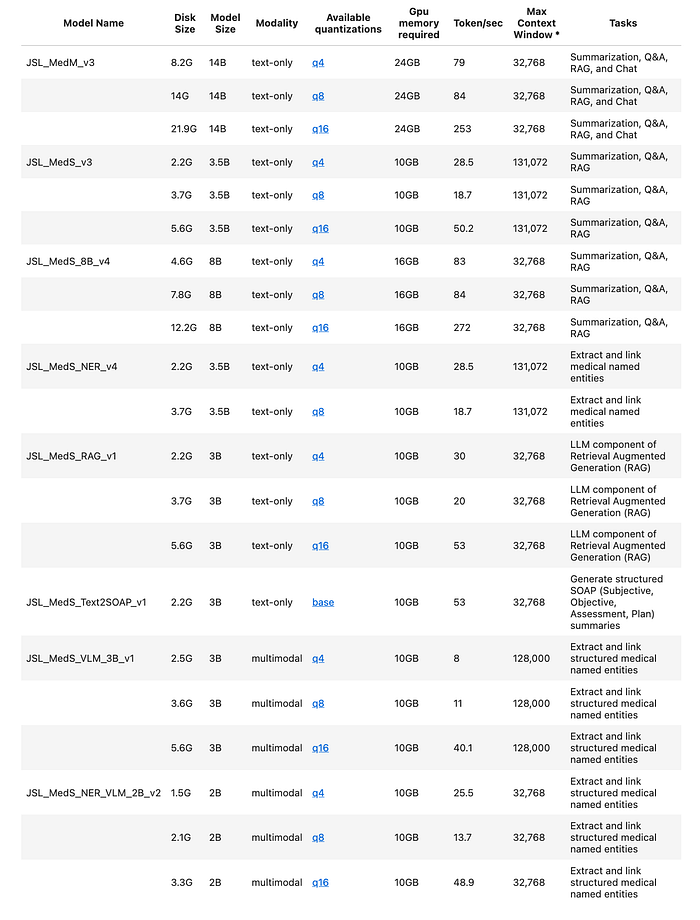
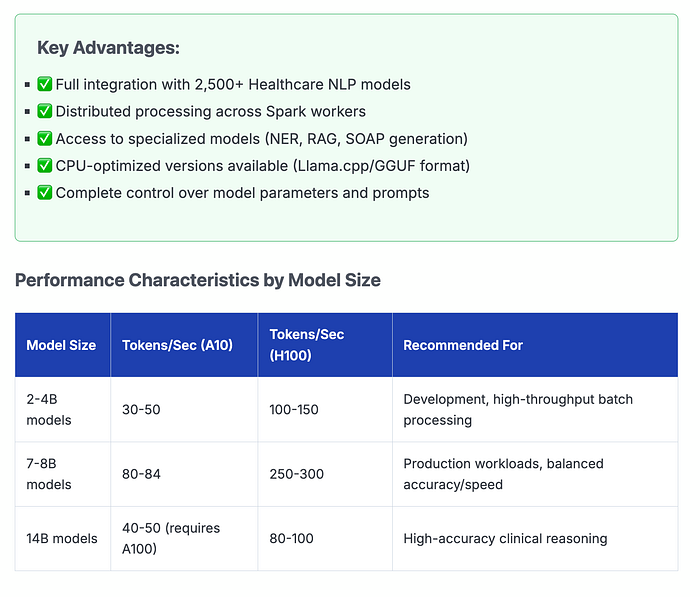
The Healthcare NLP Library is a powerful component of John Snow Labs, designed to facilitate NLP tasks within the healthcare domain. This library provides over 2,500 pre-trained models and pipelines tailored for medical data, enabling accurate information extraction, NER for clinical and medical concepts, and text analysis capabilities. Regularly updated and built with cutting-edge algorithms, the Healthcare library aims to streamline information processing and empower healthcare professionals with deeper insights from unstructured medical data sources, such as electronic health records, clinical notes, and biomedical literature.
John Snow Labs has created custom large language models (LLMs) tailored for diverse healthcare use cases. These models come in different sizes and quantization levels, designed to handle tasks such as summarizing medical notes, answering questions, performing retrieval-augmented generation (RAG), named entity recognition and facilitating healthcare-related chats.
Best for: Development, testing, and production workloads where you need maximum flexibility and integration with existing Spark NLP pipelines. See the documentation here:
This is our native approach where medical LLMs run directly within your Databricks notebooks using the Healthcare NLP library. You get access to our full suite of models across various sizes.
from johnsnowlabs import nlp, medical
# Initialize Spark session with your license
spark = nlp.start()
# Load document assembler
document_assembler = nlp.DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
# Load medical LLM
# Recommended: 8B q8 model for A10 GPU (24GB VRAM)
medical_llm = medical.AutoGGUFModel.pretrained("jsl_meds_8b_q8_v4", "en", "clinical/models")\
.setInputCols("document")\
.setOutputCol("completions")\
.setBatchSize(1)\
.setNPredict(500)\
.setUseChatTemplate(True)\
.setTemperature(0)
# Create pipeline
pipeline = nlp.Pipeline(stages=[
document_assembler,
medical_llm
])
# Process clinical data
results = pipeline.fit(data).transform(data)
Option 2: Databricks Container Services
Best for: Teams needing containerized deployments with GPU acceleration and reproducible environments.
This option uses a pre-built Docker container that you deploy on your Databricks cluster. The container includes all necessary dependencies, optimized libraries, and model artifacts.
# Models are pre-loaded in the container
"""
available_models
|model_name | model_type | availibility|
|-----------------------|------------|-----------|
| jsl_meds_4b | text only | available |
| jsl_meds_8b | text only | available |
| jsl_medm_14b | text only | available |
| jsl_meds_vl_3b | vision | available |
| jsl_meds_vl_7b | vision | available |
| jsl_meds_vl_4b | vision | will be available |
| jsl_meds_vl_8b | vision | will be available |
| jsl_medm_vl_30b | vision | available |
| jsl_meds_reasoning_8b | reasoning | available |
| jsl_medm_reasoning_32b| reasoning | available |
"""
from jsl_llm_lib.jsl_llm import JslLlm
llm = JslLlm()
llm.load_model(llm_name="jsl_meds_4b")
text = """Climate change refers to long-term shifts in temperatures and weather patterns. These shifts may be natural, but since the 1800s, human activities have been the main driver, primarily due to burning fossil fuels like coal, oil, and gas. Burning these materials releases greenhouse gases, which trap the sun’s heat and raise global temperatures. Consequences include rising sea levels, more extreme weather events, and disruptions to food and water supply. Addressing climate change requires global cooperation to reduce emissions, transition to renewable energy sources, and implement sustainable practices across various sectors."""
prompt = f"""You are a **summarization system**.
### Instruction:
You will be given a long piece of text. Summarize it into a **short version that is no more than 30–40% of the original length**.
### Text:
{text}
### Response:
"""
result = llm.get_prediction(prompt=prompt)
print("Result:",f"{result}")
'''
Result: Climate change, driven mainly by human activities since the 1800s—especially fossil fuel burning—has led to increased greenhouse gas emissions, rising global temperatures, and severe consequences like sea level rise, extreme weather, and disrupted resources. Mitigation requires global efforts to cut emissions, shift to renewable energy, and adopt sustainable practices.
'''
}
MultiModal (vision) Models
from jsl_llm_lib.jsl_llm import JslLlm
import os
llm = JslLlm()
llm.load_model(llm_name="jsl_meds_vl_3b")
images_dir = "images"
os.makedirs(images_dir, exist_ok=True)
os.system(f"wget -O {images_dir}/prescription.png -q \"https://raw.githubusercontent.com/JohnSnowLabs/spark-nlp-workshop/master/healthcare-nlp/data/ocr/prescription_02.png\"")
image_path = os.path.join(images_dir, "prescription.png")
prompt = """Extract demographic, clinical disease and medication informations"""
result = llm.get_prediction(prompt=prompt, image_path=image_path)
print("Result:",f"{result}")
"""
Result: **Demographic Information:**
- **Name:** Ms. Rukhsana Shaheen
- **Age:** 56 years
- **Sex:** Female
**Clinical Disease Information:**
- **Systemic Lupus Erythematosus (SLE) and Scleroderma Overlap with Interstitial Lung Disease (ILD)**
- **Present problem:** Tightness of skin of the fists and ulcers on the pulp of the fingers
**Medication Information:**
- **Linezolid 600 mg twice a day for 5 days** (as advised in detail in case the ulcers of the fingers do not heal)
- **Clopidogrel 75 mg once a day after meals**
- **Amlodipine 5 mg once a day**
- **Domperidone 10 mg twice a day before meals**
- **Omeprazole 20 mg twice a day before meal**
- **Bosentan 62.5 mg twice a day after meals**
- **Sildenafil Citrate 0.5 mg twice a day after meals**
- **Prednisolone 5 mg once a day after breakfast**
- **Mycophenolate mofetil 500 mg 2 tablets twice a day**
- **L-methylfolate calcium 400 μg 1 tablet once a day**
- **Ciprofloxacin 250 mg twice a day**
- **Review after 4 weeks**
"""
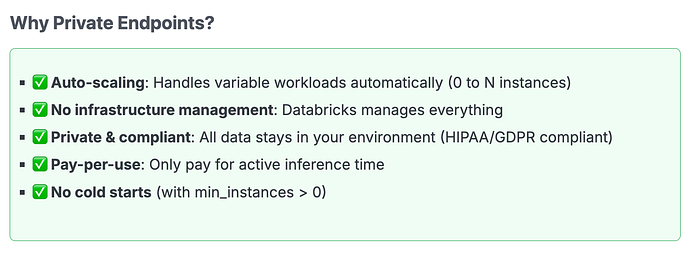
Option 3: Databricks Private Endpoints
Best for: Production deployments requiring auto-scaling, minimal infrastructure management, and API-based access.
Private endpoints provide a fully managed service where Databricks handles infrastructure while keeping your data and models within your private network. This is our recommended approach for production at scale.
https://learn.microsoft.com/en-us/azure/databricks/partners/ml/john-snow-labs
import requests
import json
# Your private endpoint URL (provided after subscription)
endpoint_url = "https://.databricks.com/serving-endpoints/jsl-medical-llm-32b"
headers = {
"Authorization": f"Bearer {databricks_token}",
"Content-Type": "application/json"
}
# Clinical summarization request
payload = {
"inputs": {
"prompt": """
Summarize the following clinical note into a concise assessment:
[Clinical note text here...]
""",
"max_tokens": 500,
"temperature": 0.0
}
}
# Make synchronous call
response = requests.post(endpoint_url, headers=headers, json=payload)
result = response.json()
print(result["completions"][0]["text"])
{
"min_instances": 0, # Scale to zero when idle
"max_instances": 5,
"scale_down_delay": "5m"
}
# Pros: Minimal cost during idle periods
# Cons: ~30-60s cold start on first request
{
"min_instances": 2, # Always warm
"max_instances": 20,
"target_utilization": "60%",
"scale_down_delay": "15m"
}
# Pros: No cold starts, smooth scaling
# Cons: Higher minimum cost
Choose Healthcare NLP Library if:
- You need to combine LLMs with specialized NER/RE models
- Building complex multi-stage pipelines (e.g., NER → RAG → Summarization)
- Want maximum flexibility in prompt engineering and model chaining
- Already using Spark NLP for other healthcare tasks
Choose Container Services if:
- Standardizing deployments across multiple teams
- Need reproducible environments for compliance/audit
- Prefer Docker-based MLOps workflows
- Want to version-lock your entire stack
Choose Private Endpoints if:
- Processing variable workloads (e.g., batch jobs during specific hours)
- Building microservices or REST APIs consuming LLMs
- Want to minimize DevOps overhead
- Need auto-scaling for unpredictable traffic patterns
Key Takeaways
- John Snow Labs provides the most accurate medical LLMs on the market, outperforming GPT-4o, Claude, and Med-PaLM-2 on healthcare benchmarks by 4–11 percentage points
- Three flexible deployment options on Databricks let you balance control, scalability, and ease of use based on your specific requirements
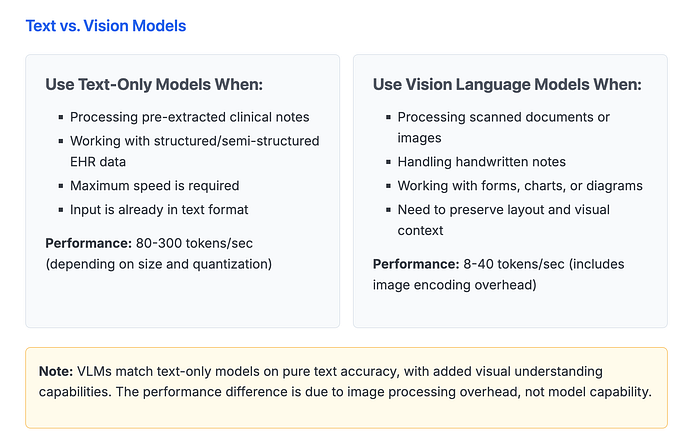
- Vision Language Models (VLMs) can process complex medical documents, including handwritten notes and scanned images, with accuracy preferred by medical practitioners 46–175% more often than GPT-4o
- All deployments are HIPAA/GDPR compliant with no data leaving your secure environment — a critical advantage over cloud-based APIs
- Model sizes range from 2B to 70B parameters, allowing you to choose the right balance of accuracy and speed for your GPU resources and budget
- Specialized medical models (NER, RAG, SOAP generation) provide domain-specific capabilities not available in general-purpose LLMs
- Production-ready performance with 80–300 tokens/sec on modern GPUs and auto-scaling capabilities for variable workloads
Complete Notebooks:
- Loading Medical LLMs (Text-Only Models)
- Multi-Modal Vision Language Models
- Medical LLM on Databricks
📖 Documentation: