In this post, we explore the utilization of pre-trained models within the Healthcare NLP library by John Snow Labs to map medical terminology to the MedDRA ontology. Specifically, our aim is to facilitate standardized categorization for enhanced medical data analysis and interpretation.
Let us start with a short Spark NLP introduction and then discuss the details of the response to cancer treatment with some solid results.
Spark NLP & LLM in Healthcare
The Healthcare Library is a powerful component of John Snow Labs’ Spark NLP platform, designed to facilitate NLP tasks within the healthcare domain. This library provides over 2,200 pre-trained models and pipelines tailored for medical data, enabling accurate information extraction, Named Entity Recognition (NER) for clinical and medical concepts, and text analysis capabilities. Regularly updated and built with cutting-edge algorithms, the Healthcare library aims to streamline information processing and empower healthcare professionals with deeper insights from unstructured medical data sources, such as electronic health records, clinical notes, and biomedical literature.
John Snow Labs’ GitHub repository serves as a collaborative platform where users can access open-source resources, including code samples, tutorials, and projects, to further enhance their understanding and utilization of Spark NLP and related tools.
John Snow Labs also offers periodic certification trainings to help users gain expertise in utilizing the Healthcare Library and other components of their NLP platform.
John Snow Labs’ demo page provides a user-friendly interface for exploring the capabilities of the library, allowing users to interactively test and visualize various functionalities and models, facilitating a deeper understanding of how these tools can be applied to real-world scenarios in healthcare and other domains.
What is MedDRA(Medical Dictionary for Regulatory Activities)?
MedDRA is a clinically-validated international medical terminology used by regulatory authorities and the regulated biopharmaceutical industry. The terminology is used through the entire regulatory process, from pre-marketing to post-marketing, and for data entry, retrieval, evaluation, and presentation.
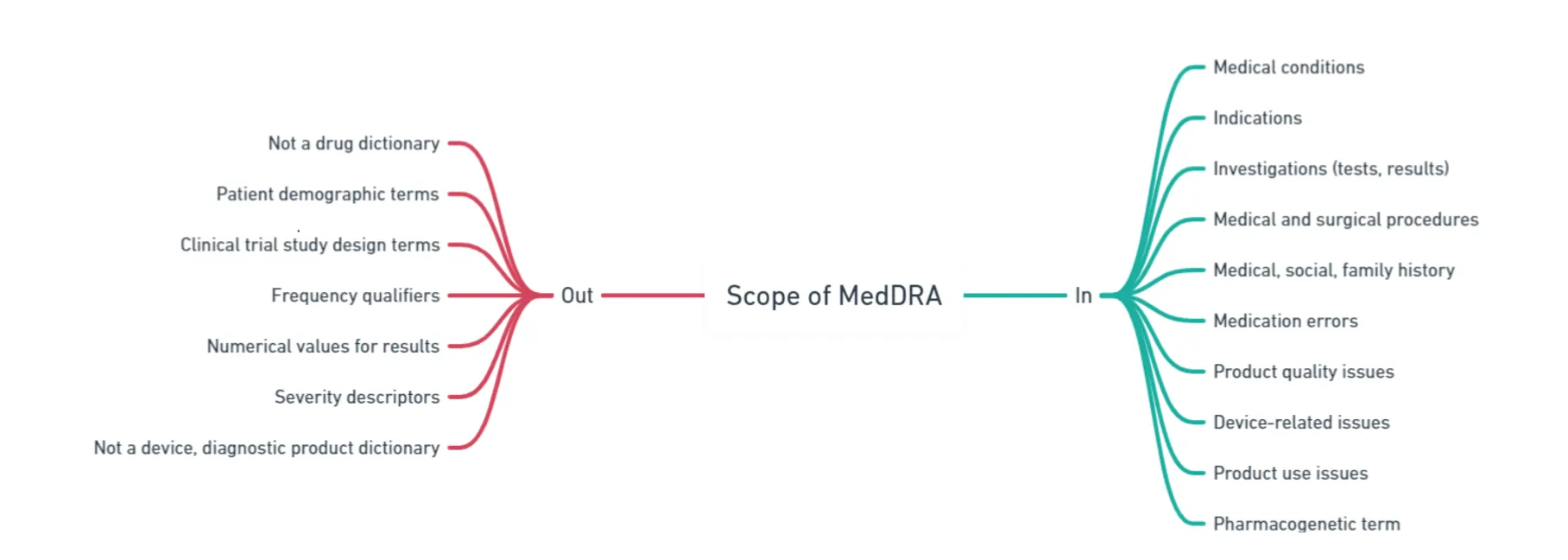
MedDRA is structured hierarchically;
System Organ Classes (SOCs): SOCs are general categories that represent different body systems or medical areas. Some examples of SOCs are: Heart problems (Cardiac disorders), Nervous system issues (Nervous system disorders) In total, there are 27 SOCs.
High-Level Group Terms (HLGTs): Within each SOC are HLGTs. HLGTs further specify the categories within a SOC. They group similar medical conditions or diseases. For example, within the SOC “Nervous system disorders,” there could be an HLGT named “Headache and migraine disorders.”
High-Level Terms (HLTs): HLTs further refine high-level group terms (HLGTs) by providing more specific descriptions of medical conditions. They still represent broader groups of related terms but with greater detail. For example, the HLT “Migraine disorders” falls under the HLGT “Headache and migraine disorders.”
Preferred Terms (PTs): PTs are the most specific level in the MedDRA hierarchy. They represent individual medical concepts or terms. PTs provide highly detailed descriptions of specific medical events or conditions. Within the HLT “Migraine disorders,” for instance, a PT could be “Migraine with aura,” which indicates the specific type of migraine.
Lowest Level Terms (LLTs): LLTs are equivalent to Preferred Terms (PTs) and represent the most specific and detailed descriptions of medical concepts.
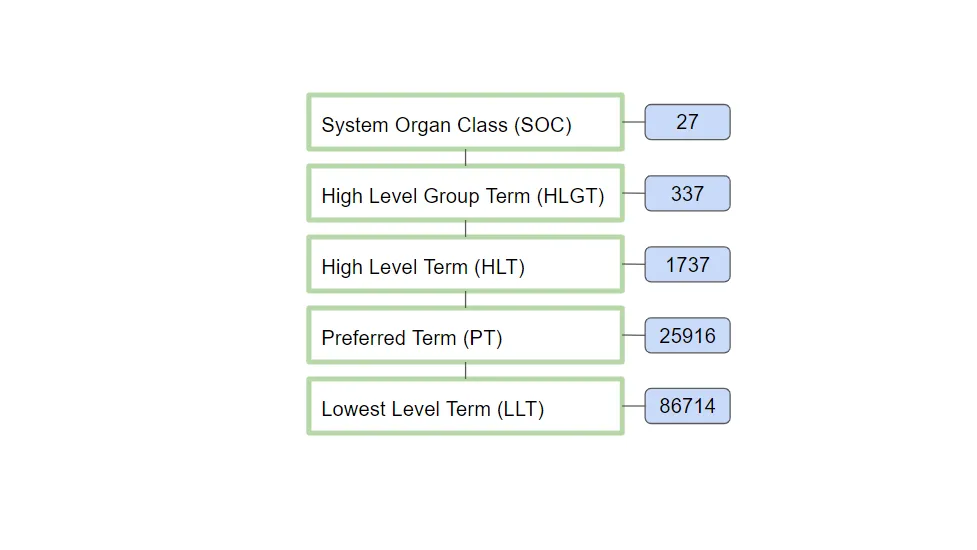
MedDRA Structure
As you navigate the medical dictionary in more depth, the terms become more specific. Preferred Terms (PTs) are the most specific and exact medical terms, providing a clear foundation for coding and communication in the healthcare field.
Detecting and Mapping MedDRA Concepts in Free-Text Documents
In Spark NLP for Healthcare, the process of mapping entities to medical terminologies, or entity resolution, begins with Named Entity Recognition (NER). First we need to extract the related clinical entities from clinical texts by using clinical NER models in Spark NLP. After getting the appropriate entities, we feed these entity chunks to the Sentence BERT (SBERT) stage, which generates embeddings for each entity. These embeddings are then fed into the entity resolution stage, which utilizes a pre-trained model to return the closest terminology code based on semantic similarity measures between the named entity (chunk) and descriptions within the medical terminology database. It is basically finding the most similar concepts/descriptions via vector database.
Healthcare NLP comes with 100+ different entity resolver models to support several clinical terminologies (RxNorm, ICD-10-CM, SNOMED, CPT, ATC, HPO, etc.).
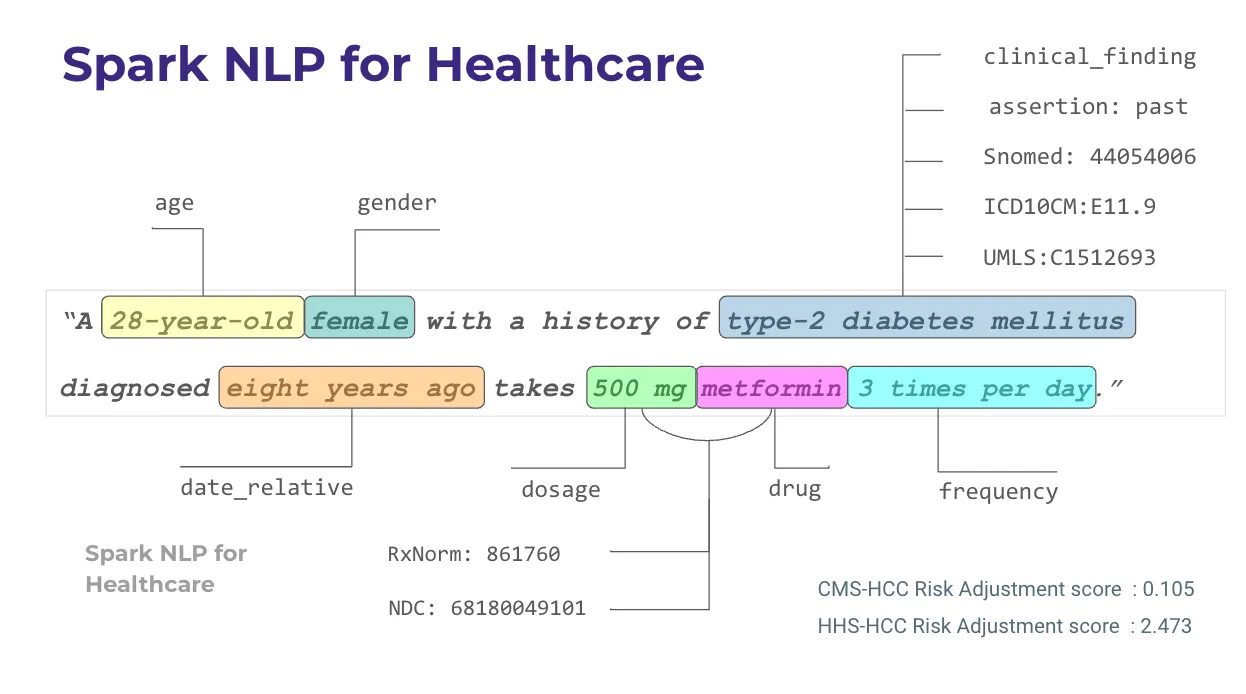
And now, new sbiobertresolve_meddra_preferred_term and sbiobertresolve_meddra_lowest_level_term models are released in v5.3.1 that map clinical terms to MedDRA codes.
| Model Name | Description |
|---|---|
| sbiobertresolve_meddra_lowest_level_term | This model maps clinical terms to their corresponding MedDRA LLT (Lowest Level Term) codes. |
| sbiobertresolve_meddra_preferred_term | This model maps clinical terms to their corresponding MedDRA PT (Preferred Term) codes. |
| icd10_meddra_llt_mapper | Maps ICD-10 codes to corresponding MedDRA LLT (Lowest Level Term) codes. |
| meddra_llt_icd10_mapper | Maps MedDRA-LLT (Lowest Level Term) codes to corresponding ICD-10 codes. |
| icd10_meddra_pt_mapper | Maps ICD-10 codes to corresponding MedDRA-PT (Preferred Term) codes. |
| meddra_pt_icd10_mapper | Maps MedDRA-PT (Preferred Term) codes to corresponding ICD-10 codes. |
| meddra_llt_pt_mapper | Maps MedDRA-LLT (Lowest Level Term) codes to their corresponding MedDRA-PT (Preferred Term) codes. |
| meddra_pt_llt_mapper | Maps MedDRA-PT (Preferred Term) codes to their corresponding MedDRA-LLT (Lowest Level Term) codes. |
You can find an example of these models below. Also, for an extended example of usage and to see the full pipeline, refer to the MedDRA notebook in the Spark NLP Workshop repository.
meddra_resolver = SentenceEntityResolverModel.load("sbiobertresolve_meddra_preferred_term") \
.setInputCols(["sbert_embeddings"]) \
.setOutputCol("meddra_pt_code")\
.setDistanceFunction("EUCLIDEAN")
Result:
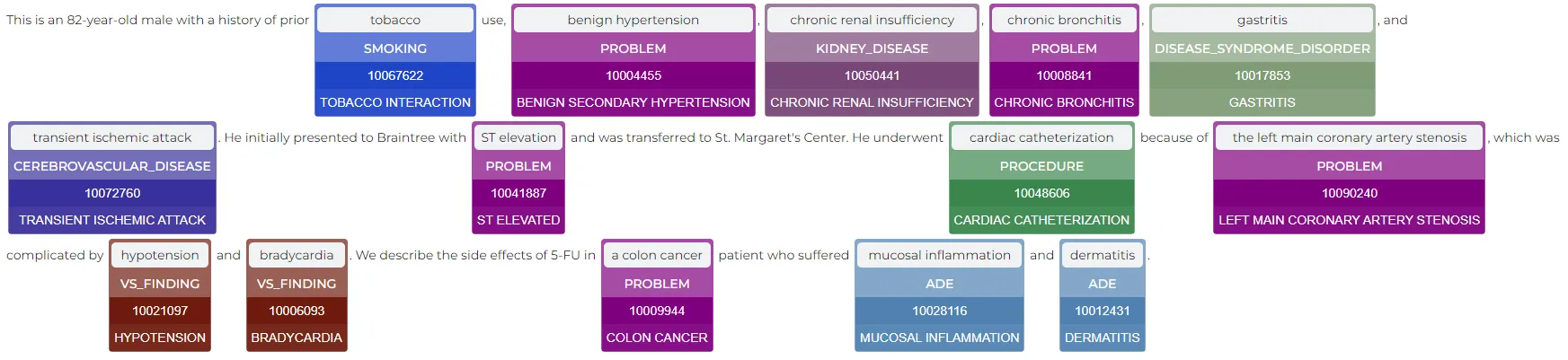
Automated Mappings between MedDRA and ICD-10
In Spark NLP, there are 60+ chunk mapper models that were trained for several solutions like mapping clinical terminology codes interchangeably. Detailed examples can be found in the Chunk Mapping Notebook.
In the latest release, v5.3.0, Spark NLP introduces four additional chunk mapper models, further expanding its collection alongside the existing 60+ models.
meddra_pt_icd10_mapper model maps MedDRA-PT (Preferred Term) codes to corresponding ICD10 codes.
mapperModel = ChunkMapperModel.load("meddra_pt_icd10_mapper")\
.setInputCols(["meddra_code"])\
.setOutputCol("mappings")\
.setRels(["icd10_code"])
Result:
+-----------+-------------------------------------+------------------------------------------------------------------------------------------------------------------------------------------+ |meddra_code|icd10_code |all_k_resolutions | +-----------+-------------------------------------+------------------------------------------------------------------------------------------------------------------------------------------+ |10000153.0 |O62:Abnormalities of forces of labour|O62:Abnormalities of forces of labour:::O62.8:Other abnormalities of forces of labour:::O62.9:Abnormality of forces of labour, unspecified| |10000081.0 |R10:Abdominal and pelvic pain |R10:Abdominal and pelvic pain:::R10.4:Other and unspecified abdominal pain | |10039085.0 |J30:Vasomotor and allergic rhinitis |J30:Vasomotor and allergic rhinitis:::J30.3:Other allergic rhinitis:::J30.4:Allergic rhinitis, unspecified | +-----------+-------------------------------------+------------------------------------------------------------------------------------------------------------------------------------------+
meddra_llt_icd10_mapper model maps MedDRA-LLT (Lowest Level Term) codes to corresponding ICD10 codes.
mapperModel = ChunkMapperModel.load('meddra_llt_icd10_mapper')\
.setInputCols(["meddra_code"])\
.setOutputCol("mappings")\
.setRels(["icd10_code"])
Result:
+-----------+-------------------------------------------------+--------------------------------------------------------+ |meddra_code|icd10_code |all_k_resolutions | +-----------+-------------------------------------------------+--------------------------------------------------------+ |10045275 |A01:Typhoid and paratyphoid fevers |A01:Typhoid and paratyphoid fevers:::A01.0:Typhoid fever| |10067585 |E11:Type 2 diabetes mellitus |E11:Type 2 diabetes mellitus::: | |10026182 |C15.9:Malignant neoplasm: Oesophagus, unspecified|C15.9:Malignant neoplasm: Oesophagus, unspecified::: | +-----------+-------------------------------------------------+--------------------------------------------------------+
We also possess inverse models capable of mapping ICD-10 codes to Preferred Terms (PTs) along with their corresponding LLT codes.
Easy-to-use, Out-of-the-box MedDRA Pipelines
meddra_pt_resolver_pipeline: This dedicated pipeline extracts clinical terms and utilizes sbiobert_base_cased_mli Sentence Bert Embeddings to link them to their corresponding MedDRA PT (Preferred Term) codes. Additionally, the pipeline converts MedDRA PT codes into MedDRA Lowest Level Term (LLT) codes with the meddra_pt_llt_mapper model and into ICD-10 codes using the meddra_pt_icd10_mapper model.
from sparknlp.pretrained import PretrainedPipeline
meddra_pt_pipeline = PretrainedPipeline.from_disk("meddra_pt_resolver_pipeline")
result = meddra_pt_pipeline.fullAnnotate("""I've been using Arthrotec 50 on and off for over 10 years, only taking it when necessary. However, I have begun experiencing dizziness and tinnitus due to this medication. Due to worsening arthritis, I started taking it twice a day at age 50, which led to gastritis.""")
Result:
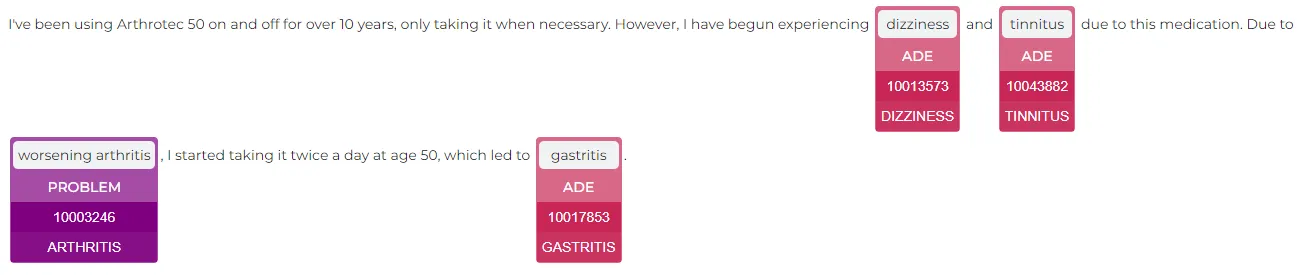
sbiobert_base_cased_mli Sentence Bert Embeddings to link them to their corresponding MedDRA LLT (Lowest Level Term) codes. Additionally, the pipeline converts MedDRA LLT codes into MedDRA Preferred Term (PT) codes with the meddra_llt_pt_mapper model and into ICD-10 codes using the meddra_llt_icd10_mapper model.
from sparknlp.pretrained import PretrainedPipeline
meddra_llt_pipeline = PretrainedPipeline.from_disk("meddra_llt_resolver_pipeline")
result = meddra_llt_pipeline.fullAnnotate('We describe the side effects of 5-FU in a colon cancer patient who suffered nausea, dermatitis, prolonged myelosuppression, and neurologic toxicity that required admission to the intensive care unit. Anterior lumbosacral radiculopathy after intrathecal methotrexate treatment and acute erythroid leukemia after cyclophosphamide therapy for plasma cell myeloma: report of two cases.')
Result:
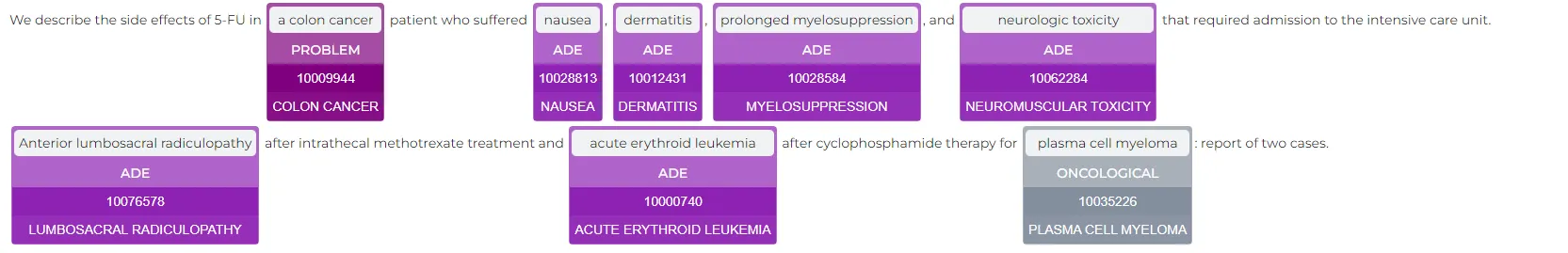
Conclusion
The utilization of Healthcare NLP in mapping medical terms to the MedDRA ontology presents a significant advancement in the field of regulatory affairs. The hierarchical structure of MedDRA, from System Organ Classes to Preferred Terms, allows for granular categorization and precise identification of medical concepts, enhancing the efficiency and accuracy of regulatory activities.
Recent progress in healthcare NLP shows a shift toward real-time ontology mapping integrated directly into hospital information systems. Deployments tested across large hospital networks in 2025 demonstrated that automated MedDRA–ICD alignment not only accelerates regulatory reporting but also supports predictive safety monitoring, allowing clinicians to identify adverse drug reactions before they escalate. This proactive use of NLP pipelines highlights their growing role in patient safety initiatives.
Another key trend is the integration of multimodal data streams into ontology mapping. Clinical notes are now increasingly combined with imaging findings, lab results, and even wearable device data. In trials conducted by academic medical centers last year, NLP models enriched with multimodal signals achieved higher precision in mapping complex oncology cases to MedDRA categories, which in turn improved outcomes in post-treatment follow-up and pharmacovigilance.
Finally, explainable NLP has become a priority for both healthcare providers and regulators. Newer versions of entity resolution models now include transparent scoring systems that show why a particular MedDRA code was selected. Pilot implementations in European regulatory agencies during 2025 revealed that this interpretability not only improves trust in AI-assisted workflows but also speeds up audits by providing human reviewers with clear evidence trails.
Furthermore, the integration of automated mappings between MedDRA and ICD-10 facilitates interoperability between different medical coding systems, promoting standardized data exchange and enhancing communication across healthcare domains.
With the availability of easy-to-use, out-of-the-box MedDRA pipelines, organizations can seamlessly incorporate MedDRA terminology into their workflows, reducing manual effort and ensuring compliance with regulatory requirements.
Overall, the adoption of Healthcare NLP for mapping medical terms to MedDRA not only improves the regulatory process but also contributes to better patient safety and healthcare outcomes by enabling comprehensive analysis and understanding of medical data.
FAQ
1) What problem does MedDRA mapping with Healthcare NLP actually solve?
It standardizes free-text clinical narratives (e.g., adverse event notes) into MedDRA codes so data can be searched, analyzed, audited, and reported consistently. This reduces manual coding effort, shortens regulatory turnaround times, and improves pharmacovigilance signal detection.
2) How does Spark NLP for Healthcare perform MedDRA mapping end-to-end?
Pipelines extract spans with clinical NER, generate embeddings (SBERT), and resolve each span to the closest MedDRA concept using pretrained entity resolvers. Optional chunk mappers translate between MedDRA and ICD-10 to support downstream analytics and reporting.
3) When should I use PT vs. LLT in MedDRA?
Use Preferred Terms (PTs) for analysis and reporting because they’re the canonical concepts. Use Lowest Level Terms (LLTs) to capture the exact wording seen in the source text and to preserve clinical nuance; LLTs can be programmatically rolled up to their PTs.
4) How do teams ensure accuracy and trust in automated coding?
Combine high-quality pretrained models with human-in-the-loop review for edge cases, and log resolver confidence scores plus alternative candidates. Track coding drift over time, add feedback loops to retrain or update dictionaries, and keep a clear audit trail for reviewers.
5) Can this integrate with EHRs and regulatory workflows (e.g., safety reports)?
Yes. The pipelines can run batch or streaming, accept HL7/FHIR inputs, and emit MedDRA/ICD-10 outputs for safety systems and dashboards. Role-based access, de-identification, and PHI minimization should be applied to meet privacy and compliance requirements.