In the world of natural language processing, the ability to extract meaningful relationships between entities within text data is crucial for various applications. This blog post will delve into three powerful features that enhance Relation Extraction: “Filter By Token Distance”, “Direction Sensitive” and “Scope Window.”
Spark NLP & LLM
The Healthcare Library is a powerful component of John Snow Labs’ Spark NLP platform, designed to facilitate NLP tasks within the healthcare domain. This library provides over 2,200 pre-trained models and pipelines tailored for medical data, enabling accurate information extraction, NER for clinical and medical concepts, and text analysis capabilities. Regularly updated and built with cutting-edge algorithms, the Healthcare library aims to streamline information processing and empower healthcare professionals with deeper insights from unstructured medical data sources, such as electronic health records, clinical notes, and biomedical literature.
John Snow Labs’ GitHub repository serves as a collaborative platform where users can access open-source resources, including code samples, tutorials, and projects, to further enhance their understanding and utilization of Spark NLP and related tools.
John Snow Labs also offers periodic trainings to help users gain expertise in utilizing the Healthcare Library and other components of their NLP platform.
John Snow Labs’ demo page provides a user-friendly interface for exploring the capabilities of the library, allowing users to interactively test and visualize various functionalities and models, facilitating a deeper understanding of how these tools can be applied to real-world scenarios in healthcare and other domains.
Introduction
In the blog post titled “Connecting the Dots in Clinical NLP using Relation Extraction Models in Spark NLP”, an insightful overview of the importance of relation extraction in the clinical domain and how Spark NLP for Healthcare facilitates this process is provided.
Relation extraction involves identifying and extracting connections between named entities within text. In clinical contexts, this process holds significance as it allows for the extraction of valuable insights from medical documents, aiding in patient care enhancement, improved clinical decision-making, and facilitating clinical research. Examples of related clinical entities encompass diseases, symptoms, treatments, and medications.
Let’s explore constructing a complete pipeline to extract all necessary information in the end.
document_assambler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentence_detector = SentenceDetector()\
.setInputCols(["document"])\
.setOutputCol("sentence")
tokenizer = sparknlp.annotators.Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")
word_embeddings = WordEmbeddingsModel()\
.pretrained("embeddings_clinical", "en", "clinical/models")\
.setInputCols(["sentence", "token"])\
.setOutputCol("embeddings")
pos_tagger = PerceptronModel()\
.pretrained("pos_clinical", "en", "clinical/models") \
.setInputCols(["sentence", "token"])\
.setOutputCol("pos_tags")
clinical_ner_tagger = MedicalNerModel()\
.pretrained("ner_clinical", "en", "clinical/models")\
.setInputCols("sentence", "token", "embeddings")\
.setOutputCol("ner_tags")
ner_chunker = NerConverterInternal()\
.setInputCols(["sentence", "token", "ner_tags"])\
.setOutputCol("ner_chunk")
dependency_parser = DependencyParserModel()\
.pretrained("dependency_conllu", "en")\
.setInputCols(["sentence", "pos_tags", "token"])\
.setOutputCol("dependencies")
clinical_re_Model = RelationExtractionModel()\
.pretrained("re_clinical", "en", 'clinical/models')\
.setInputCols(["embeddings", "pos_tags", "ner_chunk", "dependencies"])\
.setOutputCol("relations")\
.setRelationPairs(["problem-test",
"problem-treatment",])
flattener = Flattener()\
.setInputCols("relations")\
.setExplodeSelectedFields({"relations": ["result as relation_result",
"metadata.entity1 as entity1",
"metadata.entity2 as entity2",
"metadata.chunk1 as chunk1",
"metadata.chunk2 as chunk2" ] })
pipeline = Pipeline(stages=[
document_assambler,
sentence_detector,
tokenizer,
word_embeddings,
pos_tagger,
clinical_ner_tagger,
ner_chunker,
dependency_parser,
clinical_re_Model,
flattener
])
sample_df = spark.createDataFrame([["obesity with a body mass index of 33.5 kg/m2 , presented with a one-week history of polyuria and vomiting."]]).toDF("text")
model = pipeline.fit(sample_df).transform(sample_df).show(truncate= False)
Output:

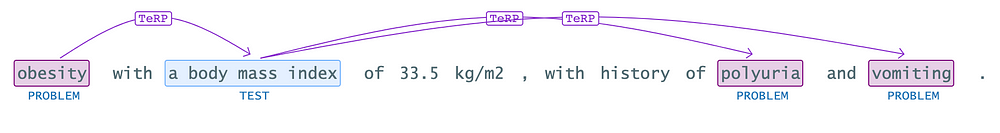
In simple terms, this relation extraction (RE) model takes named entities like “obesity,” “body mass index,” “polyuria,” and “vomiting” and identifies any relationships between them, if present.
Filtering Relations with Token Distance
The “filterByTokenDistance” parameter introduces an approach to relation extraction by considering the proximity of tokens within the text. By defining a threshold for the maximum allowable distance between tokens in a relationship, this method enables us to filter out relationships based on their spatial coherence within the text.
Here’s how it works:
- Token Distance:The method allows us to set a threshold, determining how close or far apart tokens can be while still considered part of the same relationship.
- Filtering Criteria:Relationships that meet the distance criteria are retained, while those exceeding the specified distance are filtered out.
- Optimization:Finding the optimal token distance threshold involves experimentation to maximize the accuracy of relationship extraction while minimizing noise.
By fine-tuning this parameter, we can optimize the quality of relationship extraction results, ensuring relevance and coherence in the identified connections.
clinical_re_Model = RelationExtractionModel()\
.pretrained("re_clinical", "en", 'clinical/models')\
.setInputCols(["embeddings", "pos_tags", "ner_chunk", "dependencies"])\
.setOutputCol("relations")\
.setRelationPairs(["problem-test"])
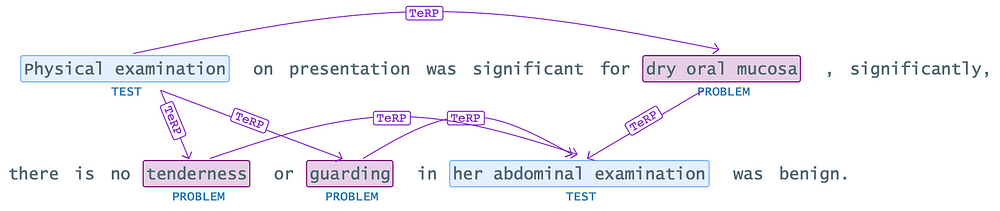
As seen in the example above, finding a relationship between ‘Physical examination’ and ‘tenderness’ or ‘guarding’ can be misleading due to deficiencies and incorrect usage of punctuation marks in the text, which can lead to such undesired results. To address such situations, we can use the ‘setFilterByTokenDistance’ method.
By utilizing the “setFilterByTokenDistance” method, we can instruct the model to consider number of tokens between chunks when identifying relationships. This allows us to filter out irrelevant or misleading relationships.
For instance, in the given example, we can set a filter to ignore relationships between “Physical examination” and symptoms like “tenderness” by specifying appropriate token distance that indicate a genuine relationship. By doing so, we can improve the accuracy of relationship extraction and avoid misleading interpretations caused by punctuation deficiencies, incorrect usage etc..
clinical_re_Model = RelationExtractionModel()\
.pretrained("re_clinical", "en", 'clinical/models')\
.setInputCols(["embeddings", "pos_tags", "ner_chunk", "dependencies"])\
.setOutputCol("relations")\
.setRelationPairs(["problem-test"])\
.setFilterByTokenDistance(8)
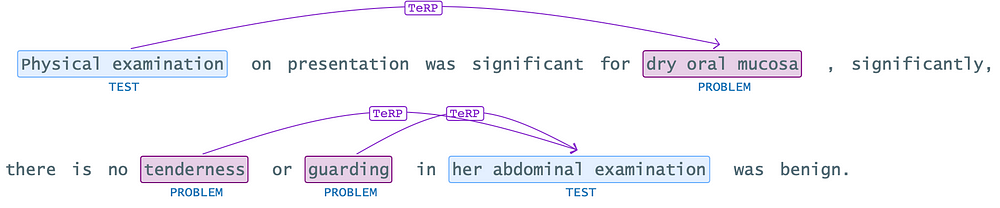
Setting the “filterByTokenDistance” to 8 offers a solution to remove the inappropriate relationship between “Physical examination” and “tenderness.” However, it’s essential to note that configuring the token distance too low might introduce unintended consequences, such as legitimate relationships not being captured. Therefore, striking a balance between filtering out irrelevant relationships and ensuring the retention of valid connections is crucial. Finding the optimal token distance threshold requires careful consideration and experimentation to achieve accurate and reliable relationship extraction results.
Direction Sensitivity for Contextual Accuracy
The “directionSensitive” parameter offers a means to discern correct relationships between entities by considering the directional context in which they appear. When enabled, only direct relations in the form of “Entity1 -> Entity2” are considered, preventing erroneous associations based solely on proximity within the text.
Here’s why it’s important:
- Contextual Accuracy:Enabling direction sensitivity ensures relationships are interpreted appropriately based on the context provided.
- Preventing Misinterpretations:It helps avoid incorrect associations that might arise from overlooking the directional flow of information within the text.
- Enhancing Analysis:By focusing on direct relationships, the accuracy of relationship extraction is enhanced, leading to more reliable insights.
re_model = RelationExtractionModel.pretrained("re_oncology_granular_wip", "en", "clinical/models")\
.setInputCols(["embeddings", "pos_tags", "ner_chunk", "dependencies"])\
.setOutputCol("relations")\
.setRelationPairsCaseSensitive(False)\
.setRelationPairs(["Biomarker-Biomarker_Result"])\
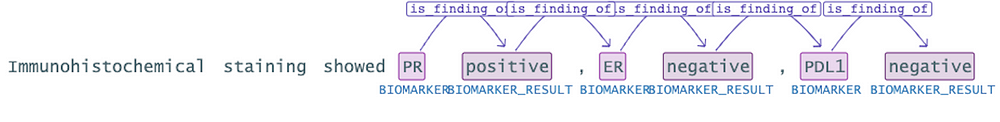
As seen in the example above, there is a relationship between the ‘PR’ and ‘positive’. However, there shouldn’t be a relationship between ‘positive’ and the ‘ER’ . The continuation of the sentence indicates that ‘ER’ is negative. To prevent such incorrect associations, it would be beneficial to utilize the “directionSensitive” parameter.
re_model = RelationExtractionModel.pretrained("re_oncology_granular_wip", "en", "clinical/models")\
.setInputCols(["embeddings", "pos_tags", "ner_chunk", "dependencies"])\
.setOutputCol("relations")\
.setRelationPairsCaseSensitive(False)\
.setRelationPairs(["Biomarker-Biomarker_Result"])\
.setDirectionSensitive(True)
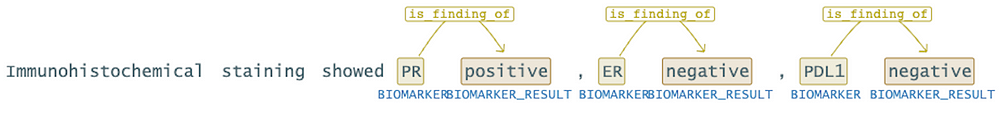
In this context, enabling the directionSensitive parameter would help in discerning the correct relationships between entities. It ensures that only direct relations in the form of “Entity1 -> Entity2” are considered, thereby avoiding erroneous associations like “positive” being linked with “ER.” This approach enhances the accuracy of the analysis and ensures that relationships are interpreted appropriately based on the context provided.
Understanding Scope Window
The Scope Window feature offers an approach to relation extraction by broadening the range of tokens. Instead of confining analysis to the immediate tokens adjacent to the entities under scrutiny, the scope window allows for the inclusion of additional tokens from the surrounding context.
clinical_re_Model = RelationExtractionModel()\
.pretrained("re_clinical", "en", 'clinical/models')\
.setInputCols(["embeddings", "pos_tags", "ner_chunk", "dependencies"])\
.setOutputCol("relations")\
.setRelationPairs(["problem-treatment"])\
.setScopeWindow([2,2])
For example, consider a sequence of tokens: x1 x2 x3 A x4 B x5 x6 x7; where A and B denote entities of interest. In a traditional scenario; A, x4 and B would be important indicators for relationship extraction. However, by applying the [2, 2] coverage window, we extend the analysis to include the x2 x3 and x7 x8 tokens, thus providing a more comprehensive context for evaluating the relationship between A and B.
The essence of the scope window feature lies in its ability to extract richer and more informative features for relation extraction. By considering a broader context, the model gains access to a wealth of linguistic cues and dependencies that may otherwise remain overlooked.
Benefits of Scope Window Feature:
- Improved Contextual Understanding:By considering a wider context, the model gains a deeper understanding of the relationships between entities, thereby facilitating more accurate predictions.
- Better Generalization:By capturing a broader range of linguistic cues, the model becomes more adept at generalizing to unseen data and handling variations in language usage.
- Increased Flexibility:The scope window feature offers flexibility in model design by allowing for customization of the context window size based on specific task requirements.
Conclusion
The integration of features like “Filter By Token Distance” ,“Direction Sensitive” and “Scope Window ” represents a significant advancement in relationship extraction techniques. By refining the selection of relationships based on token proximity and directional context, these features enhance the accuracy and relevance of extracted relationships, ultimately leading to more meaningful insights from text data.
Incorporating these features into NLP pipelines empowers users to extract nuanced relationships with greater precision, paving the way for enhanced applications in various domains, from information retrieval to sentiment analysis and beyond.




























