In this post, we will explain how to use the new Spark NLP’s DrugNormalizer pre-trained models to standardize drug names and dosage units to increase the performance of other NLP pipelines that depends on the identified entities.
Introduction
Automatic drug normalization is the process of transforming drug names and dosages into a standardized format. For example, it may be important to keep all dosages in the same unit, and drug names without abbreviation. By making the drug-related entities normalized, we can better fill the information into other machine learning pipelines and achieve better results.
Examples of normalization:
- Adalimumab 54.5 + 43.2 gm: The dosage is not standardized and there is a typo
- Agnogenic one half cup: The dosage is given in natural language (half cup)
- Interferon alpha-2b 10 million unit (1 ml) injec: The dosage is a mix of number and text, and there is an abbreviation
- Aspirin 10 meq/ 5 ml oral sol: The dosage is not in the standard unit and we have an abbreviation
To address this problem, many government agencies and private organizations make efforts to create standards and normalization for drug names, descriptions, and dosage to help practitioners to keep information more structured, ease the exchange of information and improve the accessibility to health data. Some of these initiatives are:
- RxNORM: Provides normalized names for clinical drugs and links its names to many of the drug vocabularies commonly used in pharmacy management and drug interaction software.
- SNOMED CT: The Systematized Nomenclature of Medicine – Clinical Terms, maintained by the not-for-profit association International Health Terminology Standards Development Organisation (London – UK). It contains clinical terminology for use in U.S. Federal Government systems for the electronic exchange of clinical health information.
- ICD: The International Statistical Classification of Diseases and Related Health Problems (ICD) is a standard for diagnostic classification for both clinical and research purposes endorsed by the World Health Organization (WHO), where version 10 has been cited more than 20,000 times. ICD defines the universe of diseases, disorders, injuries, and other related health conditions, listed in a comprehensive, hierarchical fashion.
These vocabularies/dictionaries can be explored online and linked to programs through APIs, but sometimes the search is not optimal due to the complex nature of natural language, and certain ways of writing the search query can help find the correct entry.
For this reason, John Snow Labs believes it to be a crucial tool to be added to the healthcare solutions the company provides, helping the clients to find the correct references faster and more accurately.
John Snow Labs’ Spark NLP
Spark NLP is an open-source NLP Python library built on top of Apache Spark from John Snow Labs. It is one of the top growing NLP production-ready solutions with support for all main tasks related to NLP in more than 46 languages.
Figure 1: Open-Source Spark NLP
Apart from the open-source library, the company maintains the licensed library specialized for healthcare solutions, SPARK NLP for Healthcare. According to the 2020 NLP Survey by Gradient Flow, from the users of NLP solutions in healthcare, Spark NLP accounts for 54%.
Figure 2: Spark NLP for Healthcare
Since its 2.7.3 release, Spark NLP for Healthcare contains NLP pre-trained models for drug names and dosage unit normalization, which can improve the performance of tasks that depend on correctly identifying drug entities.
Drug Entity Identification
Named Entity Recognition is a well-known NLP task used to extract useful information from free text. In the healthcare context, useful information may be, for example, drug name, dosage, strength, route, form, and frequency. Given the text:
“The patient is a 30-year-old female with a long history of insulin-dependent diabetes, type 2; coronary artery disease; chronic renal insufficiency; peripheral vascular disease, also secondary to diabetes; who was originally admitted to an outside hospital for what appeared to be acute paraplegia, lower extremities. She did receive a course of Bactrim for 14 days for UTI. Evidently, at some point in time, the patient was noted to develop a pressure-type wound on the sole of her left foot and left great toe. She was also noted to have a large sacral wound; this is in a similar location to her previous laminectomy, and this continues to receive daily care. The patient was transferred secondary to inability to participate in full physical and occupational therapy and continue medical management of her diabetes, the sacral decubitus, left foot pressure wound, and associated complications of diabetes.
She is given Fragmin 5000 units subcutaneously daily, Xenaderm to wounds topically b.i.d., Lantus 40 units subcutaneously at bedtime, OxyContin 30 mg p.o. q.12 h., folic acid 1 mg daily, levothyroxine 0.1 mg p.o. daily, Prevacid 30 mg daily, Avandia 4 mg daily, Norvasc 10 mg daily, Lexapro 20 mg daily, aspirin 81 mg daily, Senna 2 tablets p.o. q.a.m., Neurontin 400 mg p.o. t.i.d., Percocet 5/325 mg 2 tablets q.4 h. p.r.n., magnesium citrate 1 bottle p.o. p.r.n., sliding scale coverage insulin, Wellbutrin 100 mg p.o. daily, and Bactrim DS b.i.d.”
We can identify the following entities (from the last paragraph):
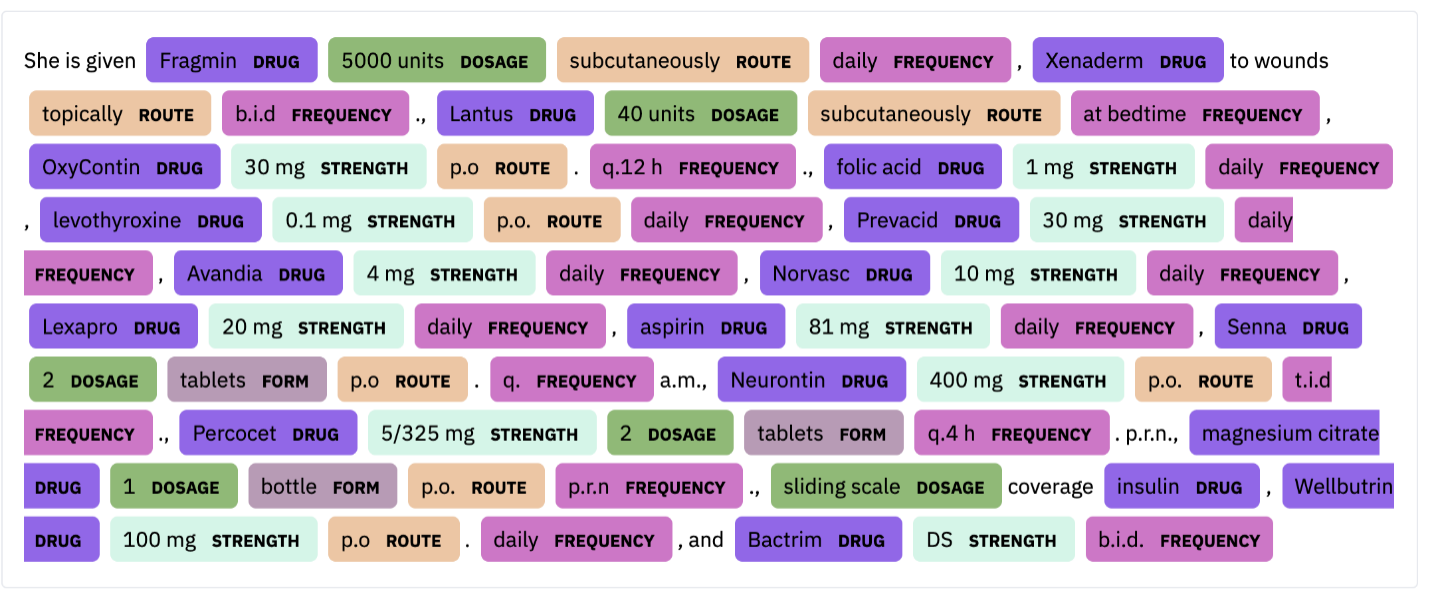
In some cases, some of the entities can be abbreviated or given in different units.
| Example | Problem(s) | Normalized |
|---|---|---|
| Adalimumab 54.5 + 43.2 gm | Dosage, typo | Adalimumab 97.7 mg |
| Agnogenic one half cup | Dosage | Agnogenic 0.5 cup |
| Interferon alpha-2b 10 million unit (1 ml) injec | Dosage, abbreviation | Interferon alpha-2b 10000000 unt (1 ml) injection |
| Aspirin 10 meq/ 5 ml oral sol | Dosage, abbreviation | aspirin 2 meq/ml oral solution |
When the entities are not normalized as in the examples from the table above, it may be harder for them to be correctly identified or to be linked to a knowledge base (entity linking, or entity resolve). In the following sections, we will exemplify how to use Spark NLP to create pipelines that improve entity resolution at scale through the new Drug Normalizer model.
Spark NLP pipeline
Spark NLP is built on top of Apache Spark and is based on the concept of pipelines, where we can perform several NLP tasks in our data with a unified pipeline. For an introduction of Spark NLP functionalities, refer to [1].
Here, we will explore the Spark NLP for Healthcare capabilities for Entity Resolution using the Drug Normalizer model. We will first divide our pipeline into two steps for clarification:
- Named Entity Resolution: Identify drug entities in text
- Entity Resolver: Link the identified drug entities to the knowledge bases
Then we will show how to use drug normalizer in these pipelines.
The NER pipeline:
# Import modules
from pyspark.ml.pipeline import Pipeline
from sparknlp.base import DocumentAssembler, LightPipeline
from sparknlp.annotator import SentenceDetectorDLModel, Tokenizer, SentenceDetector
from sparknlp_jsl.annotator import WordEmbeddingsModel, NerDLModel, NerConverter
documentAssembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentenceDetector = SentenceDetector()\
.setInputCols(["document"])\
.setOutputCol("sentence")
tokenizer = Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")
word_embeddings = WordEmbeddingsModel.pretrained("embeddings_clinical", "en", "clinical/models")\
.setInputCols(["sentence", "token"])\
.setOutputCol("embeddings")
clinical_ner = NerDLModel.pretrained("ner_clinical", "en", "clinical/models") \
.setInputCols(["sentence", "token", "embeddings"]) \
.setOutputCol("ner")
ner_converter = NerConverter() \
.setInputCols(["sentence", "token", "ner"]) \
.setOutputCol("ner_chunk")
# NER Pipeline
ner_pipeline = Pipeline(
stages = [
documentAssembler,
sentenceDetector,
tokenizer,
word_embeddings,
clinical_ner,
ner_converter])
empty_df = spark.createDataFrame([['']]).toDF('text')
ner_model = ner_pipeline.fit(empty_df)
With the NER pipeline created, we can identify related entities to be passed on a further pipeline.
The entity resolution pipeline built on top of the NER pipeline:
# Import modules
from sparknlp.base import Chunk2Doc, DocumentAssembler
from sparknlp_jsl.annotator import BertSentenceEmbeddings
from sparknlp_jsl.annotator import SentenceEntityResolverModel
c2doc = Chunk2Doc()\
.setInputCols("ner_chunk")\
.setOutputCol("ner_chunk_doc")
embeddings = BertSentenceEmbeddings\
.pretrained('sbiobert_base_cased_mli', 'en','clinical/models')\
.setInputCols(["ner_chunk"])\
.setOutputCol("sbert_embeddings")
resolver = SentenceEntityResolverModel.pretrained("sbiobertresolve_rxnorm", "en", "clinical/models") \
.setInputCols(["ner_chunk", "sbert_embeddings"]) \
.setOutputCol("rxnorm_code")\
.setDistanceFunction("EUCLIDEAN")
resolver_pipelineModel = PipelineModel(
stages = [
ner_pipeline,
c2doc,
embeddings,
resolver])
resolver_lp = LightPipeline(resolver_pipelineModel)
Here we used the stage Chunk2Doc because BertSentenceEmbeddings input columns should be an annotator of class Document, and the previous NER pipeline output was of type Chunk. The Entity Resolver pipelines will be described in more detail in another post.
Using the Drug Normalizer stage
Now, how can we use the drug normalizer in the pipeline? If we are interested in normalizing texts only, we can create a pipeline just for this:
document_assembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
drug_normalizer = DrugNormalizer() \
.setInputCols("document") \
.setOutputCol("document_normalized") \
.setPolicy( "all")
drug_normalizer_pipeline = \
Pipeline().setStages([document_assembler, drug_normalizer])
empty_df = spark.createDataFrame([['']]).toDF("text")
dn_model = drug_normalizer_pipeline.fit(empty_df)
The new DrugNormalizer stage can normalize Abbreviations, Dosages, or both (all – default), an option that can be set using the .setPolicy(policy) where policy is one of the possible values: abbreviations, dosage, or all, respectively. Additionally, it can set the option to transform the text to lower case using .setLowercase(False). The default value is False.
Now, we can use the examples in the introduction to normalize the drug names and dosage units, and then use the entity resolver to search for the entities in the RxNorm database.
First, we create a Spark data frame with the examples and normalize the entities:
# Create the Spark Data Frame
example_df = spark.createDataFrame(pd.DataFrame({'text': ["Adalimumab 54.5 + 43.2 gm",
"Agnogenic one half cup",
"Interferon alpha-2b 10 million unit (1 ml) injec",
"Aspirin 10 meq/ 5 ml oral sol"]}))
# Normalize the entities
dn_model.transform(ner_df).show()
Which gives the following output:
| text | document | sentence | token | embeddings | ner | ner_chunk | document_normalized |
|---|---|---|---|---|---|---|---|
| Adalimumab 54.5 +… | [[document, 0, 24… | [[document, 0, 24… | [[token, 0, 9, Ad… | [[word_embeddings… | [[named_entity, 0… | [[chunk, 0, 9, Ad… | [[document, 0, 18… |
| Agnogenic one hal… | [[document, 0, 21… | [[document, 0, 21… | [[token, 0, 8, Ag… | [[word_embeddings… | [[named_entity, 0… | [[chunk, 0, 21, A… | [[document, 0, 26… |
| Interferon alpha-… | [[document, 0, 47… | [[document, 0, 47… | [[token, 0, 9, In… | [[word_embeddings… | [[named_entity, 0… | [[chunk, 0, 18, I… | [[document, 0, 52… |
| Aspirin 10 meq/ 5… | [[document, 0, 28… | [[document, 0, 28… | [[token, 0, 6, As… | [[word_embeddings… | [[named_entity, 0… | [[chunk, 0, 6, As… | [[document, 0, 29… |
We then can take the output of the drug normalizer to create a new data frame with the transformed text:
normalized_df = dn_model.transform(ner_df).select(F.expr("document_normalized.result[0]").alias("text"))
normalized_df.show()
Resulting:
| text |
|---|
| Adalimumab 97700 mg |
| Agnogenic 0.5 oral solution |
| Interferon alpha – 2b 10000000 unt ( 1 ml ) injection |
| Aspirin 2 meq/ml oral solution |
Then, we can use this data frame as input to the NER pipeline and Entity Resolver as before. As an alternative, we can use the Drug Normalizer in a complete NER pipeline:
# NER stages
document_assembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentenceDetector = SentenceDetector()\
.setInputCols(["document"])\
.setOutputCol("sentence")
tokenizer = Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")
word_embeddings = WordEmbeddingsModel.pretrained("embeddings_clinical", "en", "clinical/models")\
.setInputCols(["sentence", "token"])\
.setOutputCol("embeddings")
clinical_ner = NerDLModel.pretrained("ner_posology_large", "en", "clinical/models") \
.setInputCols(["sentence", "token", "embeddings"]) \
.setOutputCol("ner")
ner_converter = NerConverter() \
.setInputCols(["sentence", "token", "ner"]) \
.setOutputCol("ner_chunk")
# Convert extracted entities to the doc with chunks in metadata
c2doc = Chunk2Doc()\
.setInputCols("ner_chunk")\
.setOutputCol("ner_chunk_doc")
drug_normalizer = DrugNormalizer() \
.setInputCols("ner_chunk_doc") \
.setOutputCol("document_normalized") \
.setPolicy( "all")
nlpPipeline = Pipeline(stages=[
document_assembler,
sentenceDetector,
tokenizer,
word_embeddings,
clinical_ner,
ner_converter,
c2doc,
drug_normalizer])
empty_data = spark.createDataFrame([[""]]).toDF("text")
model = nlpPipeline.fit(empty_data)
This complete pipeline can perform all the intermediate tasks in one run with the benefit of being able to do that at scale. The output is the normalized drug names and dosage units of the identified entities. Now, let’s see how to use the drug normalizer before identifying the entities:
document_assembler = DocumentAssembler()
.setInputCol("text")\
.setOutputCol("document")
drug_normalizer = DrugNormalizer() \
.setInputCols("document") \
.setOutputCol("document_normalized") \
.setPolicy( "all")
sentenceDetector = SentenceDetector()\
.setInputCols(["document_normalized"])\
.setOutputCol("sentence")
tokenizer = Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")
word_embeddings = WordEmbeddingsModel.pretrained("embeddings_clinical", "en", "clinical/models")\
.setInputCols(["sentence", "token"])\
.setOutputCol("embeddings")
clinical_ner = NerDLModel.pretrained("ner_posology_large", "en", "clinical/models") \
.setInputCols(["sentence", "token", "embeddings"]) \
.setOutputCol("ner")
ner_converter = NerConverter() \
.setInputCols(["sentence", "token", "ner"]) \
.setOutputCol("ner_chunk_dn")
dn_nlpPipeline = Pipeline(stages=[
document_assembler,
drug_normalizer,
sentenceDetector,
tokenizer,
word_embeddings,
clinical_ner,
ner_converter])
empty_data = spark.createDataFrame([[""]]).toDF("text")
dn_model = dn_nlpPipeline.fit(empty_data)
We added the DrugNormalizer at the beginning of the pipeline. Some times, this allows for better entity recognition, as shown in this examples (also note that in some cases it may be worse, but in general it is better):
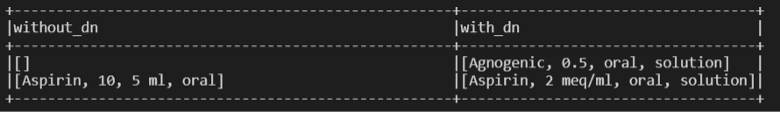
Conclusion
In this post, we introduced a new model to normalize drug names and/or dosage on clinical texts and showed how the new model improves the performance of entity resolvers in the SNOMED knowledge base. You can find an example jupyter notebook at this Colab notebook and if you want to try them on your data, you can ask for a Spark NLP Healthcare free trial license.
Being used in enterprise projects and built natively on Apache Spark and TensorFlow as well as offering all-in-one state-of-the-art NLP solutions, Spark NLP library provides simple, performant as well as accurate NLP notations for machine learning pipelines that can scale easily in a distributed environment.
If you want to find out more and start practicing Spark NLP, please check out the reference resources below.
Further reading
- Introduction to spark NLP
- Text Classification in Spark NLP
- Using BERT for NER in Spark NLP
- John Snow Labs training materials






























