PHI De-Identification with State-of-the-Art NLP
De-identification for natural language processing in healthcare is a critical procedure for safeguarding Protected Health Information (PHI) within clinical notes, wherein the data is anonymized or obfuscated through the replacement of real entities with false ones.
Multi-Mode De-identification With Healthcare NLP
We highlighted the significance of sharing clinical data in compliance with HIPAA Privacy Rules and explored the functionalities available within the Healthcare NLP library for accomplishing this objective in the blog post Format Consistency for Entity Obfuscation in De-Identification with Spark NLP.
Now we will talk about a new feature released in the latest version, v4.3.2, of Healthcare NLP (medical natural language processing) that provides the ability to obfuscate or anonymize the PHI entities in one pass.
Why Do We Need The Application of Multiple De-identification Policies At Once
There are several deidentification policies for replacing the PHI data in Healthcare NLP. Here is a list of these options;
obfuscate: Replace the values with randomly generated fake ones, eg. John Snow -> Michael Willian.mask_same_length_chars: Replace the value with the minus two same lengths asterisk and plus one bracket on both ends, eg. John Snow -> [*******].mask_entity_labels: Replace the values with the entity labels, eg. John Snow -> <NAME>.mask_fixed_length_chars: Replace the value with a fixed-length asterisk. You can also invokesetFixedMaskLength(), eg. John Snow -> ****.
The Deidentification annotator is a crucial tool within Healthcare NLP, specifically for carrying out de-identification tasks. By providing the necessary entities and specifying the desired mode of either obfuscation or mask through the setmode() parameter of this annotator, the resulting output is effectively de-identified.
In instances where a de-identified output was required using a combination of various policies, such as obfuscating both NAME and LOCATION entities while masking DATE entities with same-length characters, it became necessary to define multiple Deidentification annotators within the same pipeline for each entity label-policy pairing. Subsequently, some post-processing of the results was also required. While this requirement was very simple, the application was making the pipeline more complex and adding post-process steps was breaking the functionality of the process flow.
With the release of Spark NLP for Healthcare version 4.3.2, it is now possible to simultaneously apply multiple healthcare data de-identification policies to varying PHI entities, thanks to the newly introduced feature.
To better understand the importance of these improvements, it’s also essential to look at how the broader landscape of healthcare NLP is evolving. These technical updates are part of a much larger shift in how de-identification is applied, governed, and integrated into healthcare data workflows.
Recent advancements in healthcare NLP show a clear shift toward integrating de-identification with broader governance and compliance ecosystems. Beyond masking or obfuscating PHI, modern solutions now pair these processes with automated audit trails, traceability mechanisms, and policy-driven de-identification workflows. This allows healthcare organizations to align their NLP pipelines with regulatory frameworks like HIPAA, GDPR, and the EU AI Act without adding unnecessary manual oversight. As a result, de-identification is becoming a strategic part of secure data lifecycle management rather than a standalone technical step.
Another important trend is the adoption of adaptive de-identification powered by large language models fine-tuned on clinical and regulatory contexts. Instead of applying static masking rules, these systems dynamically adjust their policies based on context, risk level, and entity type. This enables higher precision in protecting sensitive data while maintaining the utility of clinical text for downstream applications like predictive modeling, cohort analysis, or patient journey mapping.
Finally, the industry is moving toward federated and privacy-preserving learning environments, where de-identification plays a foundational role. New architectures enable NLP models to be trained across multiple healthcare institutions without exchanging raw patient data. De-identification within these federated systems not only protects privacy but also ensures interoperability, helping organizations build more robust AI solutions without compromising compliance or security.
Implementing Multi-Mode Functionality in De-identification
We enhanced the Deidentification annotator by adding a new setSelectiveObfuscationModes() parameter which requires a JSON file that contains a user-defined dictionary with the policies which will be applied to the labels. If the entities are not provided in the JSON file, they will be deidentified according to the setMode() as default. It also provides the ability to skip entities that we don’t want to de-identify.
Let’s assume that we are working on a de-identification task and want to apply this policy combination to the document;
- obfuscate PHONE entities
- mask ID entities with entity labels
- mask NAME entities with same length chars
- mask ZIP and LOCATION entities with fixed-length chars
- DO NOT de-identify (skip) DATE entities
After creating a NER pipeline that can detect all these entities, we will define a dictionary with these policy-label pairs and save it as a JSON file. We do not need to consider the label casing while creating the dictionary, this feature is not case-sensitive for the labels. This means Zip and ZIP will return the same results.
import json
sample_json= {
"obfuscate": ["PHONE"] ,
"mask_entity_labels": ["ID"],
"skip": ["DATE"],
"mask_same_length_chars":["NAME"],
"mask_fixed_length_chars":["zip", "location"]
}
with open('multi_mode.json', 'w', encoding='utf-8') as f:
json.dump(sample_json, f, ensure_ascii=False, indent=4)
Then we will define Deidentification annotator by setting setMode('obfuscate') , and providing the path of the JSON file to setSelectiveObfuscationModes('multi_mode.json') in addition to input and output column settings. This means all the entities detected by NER models that we didn’t set specific policies will be obfuscated in the results. Also, you can invoke setFixedMaskLength() for setting the counts of fixed-length chars.
Thats all! We don’t need to define any more Deidentification annotators for the application of multi-mode de-identification policies. Let’s check how this works on a sample text:
...
deid = DeIdentification() \
.setInputCols(["sentence", "token", "ner_chunk"]) \
.setOutputCol("deidentified") \
.setMode("obfuscate")\
.setSelectiveObfuscationModesPath("sample_deid.json")\
.setFixedMaskLength(4)
text = '''
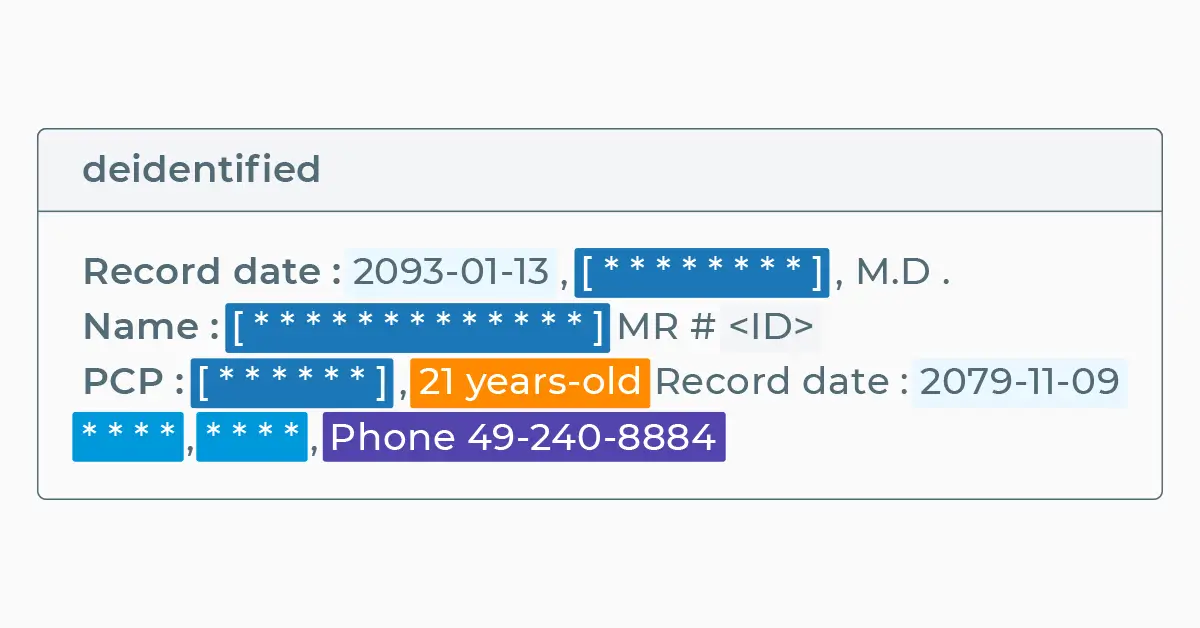
Record date : 2093-01-13 , David Hale , M.D .
Name : Hendrickson Ora MR # 7194334
PCP : Oliveira , 25 years-old Record date : 2079-11-09
Cocke County Baptist Hospital , 0295 Keats Street , Phone 55-555-5555
'''
Let’s check the results:
+---------------------------------------------------------------------+------------------------------------------------------+ |sentence |deidentified | +---------------------------------------------------------------------+------------------------------------------------------+ |Record date : 2093-01-13 , David Hale , M.D . |Record date : 2093-01-13 , [********] , M.D . | |Name : Hendrickson Ora MR # 7194334 |Name : [*************] MR # | |PCP : Oliveira , 25 years-old Record date : 2079-11-09 |PCP : [******] , 22 years-old Record date : 2079-11-09| |Cocke County Baptist Hospital , 0295 Keats Street , Phone 55-555-5555|**** , **** , Phone 97-182-9152 | +---------------------------------------------------------------------+------------------------------------------------------+
As you can see above
DATEentities were skipped:2093-01-13=>2093-01-13,2079–11–09=>2079–11–09NAMEentities were masked with same-length chars:David Hale= >[********],Hendrickson Ora=>[*************],Oliviera:[******],IDentity was masked with ID tag:7194334=><ID>AGEentity was obfuscated since we didn’t set any policy for them andset setMode()as obfuscate:25 years-old=>22 years-oldLOCATIONentities were masked with fixed-length chars:Cocke County Baptist Hospital=>****,0295 Keats Street=>****PHONEentity was obfuscated with a fake one:55-555-5555=>97-182-9152
Conclusion
Multi-mode de-identification represents an effective solution for enhancing the process flow of de-identification tasks, thanks to its functionality, integrability, and speed advantages. By leveraging this approach, it becomes possible to implement a range of obfuscation and masking policies for different entities in a streamlined manner, while also achieving faster results due to the elimination of extra pipeline stages and post-processing requirements.
De-identification is a crucial task in the NLP world and Healthcare NLP is one of the most popular libraries for this. John Snow Labs is keeping up-to-date this library with new releases every two weeks. There will be new features in the upcoming releases, keep following us!
Healthcare NLP models are licensed, so if you want to use these models, you can watch “Get a Free License For John Snow Labs NLP Libraries” video and request one from https://www.johnsnowlabs.com/install/.
You can follow us on medium and Linkedin to get further updates or join slack support channel to get instant technical support from the developers of Spark NLP. If you want to learn more about the library and start coding right away, please check our certification training notebooks.
FAQ
What is PHI de-identification in healthcare NLP?
PHI de-identification is the process of removing or obfuscating Protected Health Information from clinical text to ensure patient privacy and regulatory compliance. This is achieved through techniques such as masking, entity labeling, or generating synthetic replacements without affecting the data’s analytical value.
Why is multi-mode de-identification important?
Traditional single-mode de-identification often struggles to address different types of PHI entities with the required precision. Multi-mode de-identification allows different policies to be applied simultaneously, improving accuracy, simplifying pipeline design, and reducing post-processing steps while ensuring compliance with privacy regulations.
How does adaptive de-identification improve data security?
Unlike static masking, adaptive de-identification uses context-aware language models to adjust the level and type of de-identification dynamically. This helps preserve the utility of clinical data for research and analytics while maintaining strong privacy protection aligned with evolving regulatory standards.
Can de-identification be integrated with compliance frameworks like HIPAA and GDPR?
Yes. Modern de-identification workflows are designed to align with regulatory frameworks such as HIPAA, GDPR, and EU AI Act. They support policy-driven automation, traceability, and auditability, helping organizations meet legal requirements while securely sharing and analyzing clinical data.
What role does de-identification play in federated learning and AI innovation?
De-identification enables privacy-preserving collaboration across healthcare institutions. In federated learning environments, it allows organizations to train models on distributed data without sharing raw PHI. This fosters AI innovation while maintaining patient confidentiality and compliance with privacy regulations.