TL;DR: Working with clinical text usually means solving two problems at the same time: protecting patient privacy and keeping the information that actually matters. This article walks through new tools that is announced by John Snow Labs with Healthcare NLP 6.3.0 to help remove personal details from medical notes while still capturing useful signals like medications — without overcomplicating the process.
John Snow Labs’ latest Healthcare NLP release 6.3.0 ships a broad set of updates — new models, enhancements across existing components, and stability patches. In this section, I’m focusing on one of the most practical additions for data scientists working with clinical text: seven new pretrained NER models that help you do two things that often need to happen together in production workflows — protect privacy and retain clinical signal; which basically means keeping the medically useful content in the text after you remove PHI.
On the privacy side, the new de-identification models detect common PHI categories in messy, real-world notes, with options for either high-level labels (quick PHI coverage) or more granular PHI labels (useful for auditing and downstream logic).
The release also includes zero-shot de-ID variants, which are designed to support flexible label sets and adapt more easily to different de-identification requirements.
On the clinical extraction side, the new drug model focuses on a frequent downstream need — capturing medication mentions.
These seven new pretrained NER models cover two common needs in clinical text processing: privacy protection and extraction of drug entities. The lineup includes PHI-focused de-identification models (both generic and more granular “sub-entity” labeling), zero-shot de-ID variants (medium/large) designed for flexible label sets and faster adaptation to different requirements, and a drug entity model.
John Snow Labs de-identification and drug extraction models
Sharing patient clinical data with third parties is often necessary for research, analytics, and collaboration — but it must be done in a way that aligns with HIPAA de-identification (Privacy Rule) expectations. In practice, that means detecting and transforming PHI (Protected Health Information) so documents can be used and shared without exposing identifiers.
John Snow Labs Healthcare NLP includes a large catalog of pretrained resources for this purpose, with 260+ NER models and pipelines that support de-identification across eight languages (English, German, Spanish, Italian, French, Arabic, Romanian, and Portuguese). Beyond purely statistical NER, many pretrained de-identification pipelines also combine rule-based contextual parsers and pattern matchers to improve coverage and handle structured identifiers — helping “fill the gaps” when a neural model might miss something like a date, an ID pattern, or a contact field. In addition to de-identification, John Snow Labs also provides dedicated drug entity extraction models so teams can preserve medically relevant information — like medication mentions — after PHI has been removed.
To help users get started quickly, John Snow Labs also provides 15 dedicated de-identification notebooks that walk through common de-ID workflows and configurations. In addition, for medication extraction there is a detailed reference notebook in the GitHub repository that demonstrates how to use the drug models and interpret their outputs in practice.
Building a custom de-identification pipeline
To show how the new de-identification models can be used as building blocks (not only as full pretrained pipelines), I started with ner_deid_generic_nonMedical as a simple baseline. The goal is straightforward: run a PHI-focused NER model to detect entities such as DATE, NAME, LOCATION, ID, CONTACT, AGE, and PROFESSION, and then apply various de-identification stages to transform the original text into a privacy-safe version.
In this example, I assembled a lightweight Spark NLP pipeline that follows the standard pattern:
- Prepare text (document → sentences → tokens)
- Run embeddings + NER to produce PHI entities
- Convert entities into NER chunks
- De-identify using different masking/obfuscation strategies
This approach is useful when you want full control over the workflow — especially if you need to choose a specific NER model, plug in additional rule-based detectors, or apply a de-identification policy that matches your organization’s privacy requirements.
# Stage 1: Transforms raw text to `document`type
documentAssembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
# Stage 2: Sentence Detection/Splitting
sentenceDetector = SentenceDetectorDLModel.pretrained("sentence_detector_dl_healthcare", "en", "clinical/models")\
.setInputCols(["document"])\
.setOutputCol("sentence")
# Stage 3: Tokenization
tokenizer = Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")
# Stage 4: Clinical Embeddings
word_embeddings = WordEmbeddingsModel.pretrained("embeddings_clinical", "en", "clinical/models")\
.setInputCols(["sentence", "token"])\
.setOutputCol("embeddings")
# Step 5: NER Model
clinical_ner = MedicalNerModel.pretrained("ner_deid_generic_nonMedical", "en", "clinical/models")\
.setInputCols(["sentence", "token", "embeddings"])\
.setOutputCol("ner")
# Step 6: Converter
ner_converter = NerConverterInternal()\
.setInputCols(["sentence", "token", "ner"])\
.setOutputCol("ner_chunk")
#deid model with "entity_labels"
deid_entity_labels= DeIdentification()\
.setInputCols(["sentence", "token", "ner_chunk"])\
.setOutputCol("deid_entity_label")\
.setMode("mask")\
.setReturnEntityMappings(True)\
.setMaskingPolicy("entity_labels")
#deid model with "same_length_chars"
deid_same_length= DeIdentification()\
.setInputCols(["sentence", "token", "ner_chunk"])\
.setOutputCol("deid_same_length")\
.setMode("mask")\
.setReturnEntityMappings(True)\
.setMaskingPolicy("same_length_chars")
#deid model with "fixed_length_chars"
deid_fixed_length= DeIdentification()\
.setInputCols(["sentence", "token", "ner_chunk"])\
.setOutputCol("deid_fixed_length")\
.setMode("mask")\
.setReturnEntityMappings(True)\
.setMaskingPolicy("fixed_length_chars")\
.setFixedMaskLength(4)
#deid model with "obfuscation"
obfuscation = DeIdentification()\
.setInputCols(["sentence", "token", "ner_chunk"]) \
.setOutputCol("deidentified") \
.setMode("obfuscate")\
.setObfuscateDate(True)\
# Define the pipeline
deidPipeline = Pipeline(
stages=[
documentAssembler,
sentenceDetector,
tokenizer,
word_embeddings,
clinical_ner,
ner_converter,
deid_entity_labels,
deid_same_length,
deid_fixed_length,
obfuscation
])
# Create an empty dataframe
data = spark.createDataFrame([[""]]).toDF("text")
# Fit the dataframe to the pipeline to get the model
nlp_model = pipeline.fit(data)
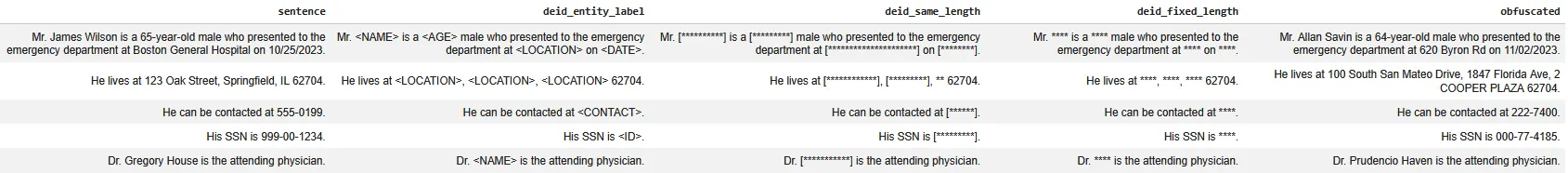
Once PHI entities are detected, the de-identification component can generate different modes (use .setMode() parameter in the Deidentification annotator) of output depending on your needs:
- Entity-label masking (
entity_labels)
Replaces detected spans with their PHI tags (e.g.,<NAME>,<DATE>). This is often the most transparent option for debugging and audits. - Same-length masking (
same_length_chars)
Masks PHI while keeping the same character length as the original span, which can help preserve formatting and alignment. - Fixed-length masking (
fixed_length_chars)
Replaces PHI spans with a constant-length mask (e.g., 4 characters). This is useful when you want uniform output and reduced leakage from token length. - Obfuscation (
obfuscate)
Substitutes PHI with realistic synthetic values (e.g., a different name or date) to keep the text readable for downstream NLP, while still removing the original identifiers.
The dataframe below show the same input text processed through these different strategies. You’ll notice that the underlying PHI detection stays consistent, but the final output changes depending on whether you want maximally explicit redaction (entity labels), format-preserving masking (same-length / fixed-length), or readable privacy-safe text (obfuscation). This makes it easy to choose the right output style based on whether your next step is manual review, dataset sharing, annotation, or downstream modeling.
Zero-shot PHI NER models
In addition to the fixed-label de-identification models, this release includes zero-shot PHI NER variants (medium/large) that can detect PHI using a user-provided label set. Instead of being limited to a predefined schema, you pass the list of entity labels you care about (e.g., MRN, EMAIL, URL, HOSPITAL, CITY, etc.), and the model attempts to find spans that match those categories. The snippet below illustrates the typical setup: standard document → sentence → token preprocessing, followed by PretrainedZeroShotNER annotator with a label list and a prediction threshold, and finally conversion to chunks for inspection/export.
document_assembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentence_detector = SentenceDetector()\
.setInputCols(["document"])\
.setOutputCol("sentence")
tokenizer = Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")
labels = ["ACCOUNTNUM", "AGE", "CITY", "COUNTRY", "DATE", "DEVICE",
"DLN", "DOCTOR", "EMAIL", "GENDER", "HOSPITAL", "IDNUM", "IP",
"LOCATION_OTHER", "MEDICALRECORD", "NAME", "ORGANIZATION", "PATIENT",
"PHONE", "PLATE", "PROFESSION", "SSN", "STATE", "STREET", "TIME", "URL",
"USERNAME", "VIN", "ZIP"]
pretrained_zero_shot_ner = PretrainedZeroShotNER().pretrained("zeroshot_ner_deid_subentity_nonMedical_medium", "en", "clinical/models")\
.setInputCols("sentence", "token")\
.setOutputCol("ner")\
.setPredictionThreshold(0.5)\
.setLabels(labels)
ner_converter = NerConverterInternal()\
.setInputCols("sentence", "token", "ner")\
.setOutputCol("ner_chunk")
pipeline = Pipeline().setStages([
document_assembler,
sentence_detector,
tokenizer,
pretrained_zero_shot_ner,
ner_converter
])
model_zero = pipeline.fit(empty_data)
Zero-shot models are particularly useful when your PHI requirements don’t match a single fixed schema, or when you want to iterate quickly across projects and datasets without retraining.
Pros
- Flexible label sets: You can add/remove labels (e.g., include
DEVICE,VIN,PLATE) without rebuilding a model. - Fast experimentation: Great for rapid prototyping — test coverage on new note types or new PHI policies immediately.
- Good “first baseline”: Helpful when you don’t yet have labeled data, but need an initial PHI layer to start processing text safely.
- Portable across domains: Can adapt to different document styles (clinical notes, intake forms, call-center text) by changing labels and thresholds.
Cons
- Less predictable than fixed-label NER: Performance can vary more with phrasing, note style, and how specific/overlapping your labels are.
- Label design matters: Too many labels, ambiguous labels, or overlapping labels (e.g.,
LOCATIONvsCITYvsSTATE) can introduce confusion. - Threshold tuning required: The prediction threshold controls sensitivity; raising it typically reduces false positives but may miss PHI, while lowering it can over-tag.
Drug entity extraction
This release includes ner_drugs_large_v2, a pretrained NER model focused on drug extraction, designed to recognize medication mentions in real-world clinical language (brand names, generics, and common shorthand).
To demonstrate usage, I built a lightweight Spark NLP pipeline that runs standard preprocessing (document → sentences → tokens), applies clinical word embeddings, and then uses ner_drugs_large_v2 to produce drug entities. The detected entities are converted into chunk annotations so they can be easily inspected, exported, or fed into downstream steps.
document_assembler = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
sentence_detector = SentenceDetector()\
.setInputCols(["document"])\
.setOutputCol("sentence")
tokenizer = Tokenizer()\
.setInputCols(["sentence"])\
.setOutputCol("token")
word_embeddings = WordEmbeddingsModel.pretrained("embeddings_clinical_large", "en", "clinical/models")\
.setInputCols(["sentence", "token"])\
.setOutputCol("embeddings")
clinical_ner = MedicalNerModel.pretrained("ner_drugs_large_v2", "en", "clinical/models") \
.setInputCols(["sentence", "token", "embeddings"]) \
.setOutputCol("ner")
ner_converter = NerConverter() \
.setInputCols(["sentence", "token", "ner"]) \
.setOutputCol("ner_chunk")
nlpPipeline = Pipeline(stages=[
document_assembler,
sentence_detector,
tokenizer,
word_embeddings,
clinical_ner,
ner_converter])
model = nlpPipeline.fit(spark.createDataFrame([[""]]).toDF("text"))
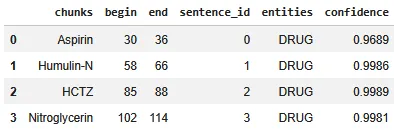
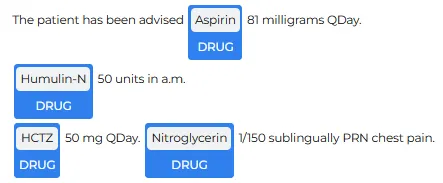
The results include a structured table of extracted spans (chunk text, character offsets, sentence id, predicted label, and confidence). In the example shown below, the model correctly identifies medication mentions such as Aspirin, Humulin N, HCTZ, and Nitroglycerin, labeling them consistently as DRUG.
The visual output highlights the same spans directly in the text, which is a quick way to validate extraction quality before moving to next stages.
Conclusion
Overall, these seven new NER models add practical building blocks for end-to-end clinical text workflows. The de-identification models help you detect PHI at either a broad level (generic labels) or with more granularity (sub-entity labels), while the zero-shot variants provide a flexible path when your labeling needs or document styles vary.
On the information extraction side, ner_drugs_large_v2 makes it easy to recover key medication signal by producing consistent Drug entities that are straightforward to export and analyze.
For data scientists, the takeaway is simple: you can assemble a privacy-first pipeline quickly—de-identify first, then extract what matters—without the overhead of building and maintaining everything from scratch.
Model Links
- ner_deid_generic_nonMedical
- ner_deid_subentity_nonMedical
- ner_drugs_large_v2
- zeroshot_ner_deid_generic_nonMedical_large
- zeroshot_ner_deid_generic_nonMedical_medium
- zeroshot_ner_deid_subentity_nonMedical_large
- zeroshot_ner_deid_subentity_nonMedical_medium