TL;DR: This blog post shows how to build an end-to-end clinical knowledge graph from unstructured medical text using John Snow Labs Healthcare NLP library. We’ll start by extracting key clinical entities about medication with Named Entity Recognition, then connect them using Relation Extraction to capture medically meaningful links. Finally, we’ll convert these structured relationships into a knowledge graph that makes complex clinical narratives easier to search, analyze, and interpret.
Clinical and biomedical data is often richest where it’s hardest to use: in free-text notes, pathology reports, discharge summaries, radiology impressions, and trial documents. These narratives contain the details that matter — diagnoses, biomarkers, treatments, disease progression, response to therapy — but they are buried in unstructured language, scattered across documents, and written with domain-specific shorthand. As a result, valuable information remains difficult to search, compare, or analyze at scale.
This is where healthcare-focused NLP becomes essential. Instead of manually reading thousands of notes, we can automatically extract clinically meaningful concepts (e.g., diseases, symptoms, drugs, tests, genetic variants, biomarker values) and convert them into structured data. But structured lists alone are not enough. In real clinical reasoning, meaning comes from connections: Which biomarker supports which diagnosis? Which drug was given for which condition? What test result triggered a treatment change?
In this blog post, we demonstrate how to turn clinical text into an interpretable, queryable knowledge graph using the John Snow Labs Healthcare NLP library. We will:
- Extract named entities using domain-trained Named Entity Recognition (NER) models
- Establish relationships using Relation Extraction models to connect entities in context
- Construct a knowledge graph that represents clinical facts as nodes and edges, enabling downstream exploration and analytics
By the end, you’ll see how this workflow transforms unstructured clinical narratives into structured, connected knowledge — making complex data more interpretable and ready for real-world applications such as cohort identification, clinical research, pharmacovigilance, and decision support.
John Snow Labs, offers a powerful NLP & LLM library tailored for healthcare, empowering professionals to extract actionable insights from medical text. Utilizing advanced techniques like NER, assertion status detection, resolver codes and relation extraction, this library helps uncover vital information for more accurate diagnosis, treatment, and prevention.
Let us start with a short Healthcare NLP introduction and then discuss the applications of John Snow Labs’ Healthcare NLP & LLM library with some solid results.
Healthcare NLP & LLM
The Healthcare Library is a powerful component of John Snow Labs’ Healthcare NLP platform, designed to facilitate NLP tasks within the healthcare domain. This library provides over 2,800 pre-trained models and pipelines tailored for medical data, enabling accurate information extraction, NER for clinical and medical concepts, and text analysis capabilities. Regularly updated and built with cutting-edge algorithms, the Healthcare library aims to streamline information processing and empower healthcare professionals with deeper insights from unstructured medical data sources, such as electronic health records, clinical notes, and biomedical literature.
John Snow Labs has created custom large language models (LLMs) tailored for diverse healthcare use cases. These models come in different sizes and quantization levels, designed to handle tasks such as summarizing medical notes, answering questions, performing retrieval-augmented generation (RAG), named entity recognition and facilitating healthcare-related chats.
John Snow Labs’ GitHub repository serves as a collaborative platform where users can access open-source resources, including code samples, tutorials, and projects, to further enhance their understanding and utilization of Healthcare NLP and related tools.
John Snow Labs also offers periodic certification training to help users gain expertise in utilizing the Healthcare Library and other components of their NLP platform.
John Snow Labs’ demo page provides a user-friendly interface for exploring the capabilities of the library, allowing users to interactively test and visualize various functionalities and models, facilitating a deeper understanding of how these tools can be applied to real-world scenarios in healthcare and other domains.
Pipeline
Let’s walk through the Healthcare NLP pipeline we use to turn raw clinical text into structured entities, relations, and graph-ready structured information.
The pipeline below will utilize the ner_posology NER model to extract medication entities from the documents, followed by the posology_re relation extraction model, which will be the foundation for knowledge graph generation.
# Step 1: Transforms raw text to `document`type
documenter = DocumentAssembler()\
.setInputCol("text")\
.setOutputCol("document")
# Step 2: Sentence Detection/Splitting
sentencer = SentenceDetector()\
.setInputCols(["document"])\
.setOutputCol("sentences")
# Step 3: Tokenization
tokenizer = sparknlp.annotators.Tokenizer()\
.setInputCols(["sentences"])\
.setOutputCol("tokens")
# Step 4: Clinical Embeddings
words_embedder = WordEmbeddingsModel()\
.pretrained("embeddings_clinical", "en", "clinical/models")\
.setInputCols(["sentences", "tokens"])\
.setOutputCol("embeddings")
# Step 5: NER Model
ner_tagger = MedicalNerModel()\
.pretrained("ner_posology", "en", "clinical/models")\
.setInputCols("sentences", "tokens", "embeddings")\
.setOutputCol("ner_tags")
# Step 6: Converter
ner_chunker = NerConverterInternal()\
.setInputCols(["sentences", "tokens", "ner_tags"])\
.setOutputCol("ner_chunks")
# Step 7: Part of speech tagger
pos_tagger = PerceptronModel()\
.pretrained("pos_clinical", "en", "clinical/models") \
.setInputCols(["sentences", "tokens"])\
.setOutputCol("pos_tags")
# Step 8: Dependency Parser
dependency_parser = DependencyParserModel()\
.pretrained("dependency_conllu", "en")\
.setInputCols(["sentences", "pos_tags", "tokens"])\
.setOutputCol("dependencies")
# Step 9: Relation Extraction
reModel = RelationExtractionModel()\
.pretrained("posology_re")\
.setInputCols(["embeddings", "pos_tags", "ner_chunks", "dependencies"])\
.setOutputCol("relations")\
.setMaxSyntacticDistance(4)
# Define the pipeline
pipeline = Pipeline(stages=[
documenter,
sentencer,
tokenizer,
words_embedder,
pos_tagger,
ner_tagger,
ner_chunker,
dependency_parser,
reModel
])
# Create an empty dataframe
data = spark.createDataFrame([[""]]).toDF("text")
# Fit the dataframe to the pipeline to get the model
nlp_model = pipeline.fit(data)
Here’s what each stage contributes and why it matters for relation extraction.
The workflow starts by converting raw clinical text into Healthcare NLP library’s internal document format and splitting it into sentences, which keeps downstream extraction grounded to sentence-level context. Each sentence is then tokenized and enriched with domain-specific clinical embeddings that capture medical semantics beyond surface wording. On top of these embeddings, a pretrained clinical NER model identifies key entities about medication—such as drugs, dosage, strength, frequency, and duration—and a converter consolidates token-level tags into span-level chunks with character offsets for traceability.
To reliably connect these entities, the pipeline adds linguistic structure via a clinical Part-of-Speech (POS) tagger and a dependency parser, producing syntactic representations of each sentence. Finally, a relation extraction model uses embeddings plus syntax (POS + dependencies) to predict clinically meaningful links between chunks (e.g., drug → strength, drug → frequency, drug → duration). The result is a set of structured triples that can be directly stored as edges in a knowledge graph, making complex clinical narratives far more searchable and interpretable.
Named Entity Recognition
NER models are crucial in identifying and categorizing entities within text, such as drugs, symptoms, medical conditions, and patient demographics. In the context of medication, the model named ner_posology can automatically recognize mentions of DOSAGE, DRUG, DURATION, FORM, FREQUENCY, ROUTE, STRENGTH from vast amounts of clinical documentation.
By systematically extracting and categorizing this information, NER models help build a structured dataset that forms the foundation for further analysis. Extracting entities in a structured format (portion of the data is shown below in json format) improves usability and integration, enabling efficient retrieval and comprehensive analysis of patient information.
[{'annotatorType': 'chunk',
'begin': 301,
'end': 308,
'result': 'five-day',
'metadata': {'sentence': '0',
'chunk': '0',
'ner_source': 'ner_chunks',
'entity': 'DURATION',
'confidence': '0.9051'}},
{'annotatorType': 'chunk',
'begin': 320,
'end': 330,
'result': 'amoxicillin',
'metadata': {'sentence': '0',
'chunk': '1',
'ner_source': 'ner_chunks',
'entity': 'DRUG',
'confidence': '0.9998'}},
{'annotatorType': 'chunk',
'begin': 378,
'end': 386,
'result': 'metformin',
'metadata': {'sentence': '1',
'chunk': '2',
'ner_source': 'ner_chunks',
'entity': 'DRUG',
'confidence': '1.0'}},
{'annotatorType': 'chunk',
'begin': 390,
'end': 398,
'result': 'glipizide',
'metadata': {'sentence': '1',
'chunk': '3',
'ner_source': 'ner_chunks',
'entity': 'DRUG',
'confidence': '0.9995'}},
{'annotatorType': 'chunk',
'begin': 406,
'end': 418,
'result': 'dapagliflozin',
'metadata': {'sentence': '1',
'chunk': '4',
'ner_source': 'ner_chunks',
'entity': 'DRUG',
'confidence': '0.9996'}},
{....
John Snow Labs’ NER Visualizer provides a user-friendly interface for visualizing the results of NER models. It highlights and categorizes identified entities within the text. This tool allows users to see how the NER models extract and label entities, making it easier to understand and interpret the extracted data. The visualizer helps in validating the accuracy of the models, identifying patterns, and gaining insights from unstructured medical data, ultimately facilitating better data analysis and decision-making in healthcare.
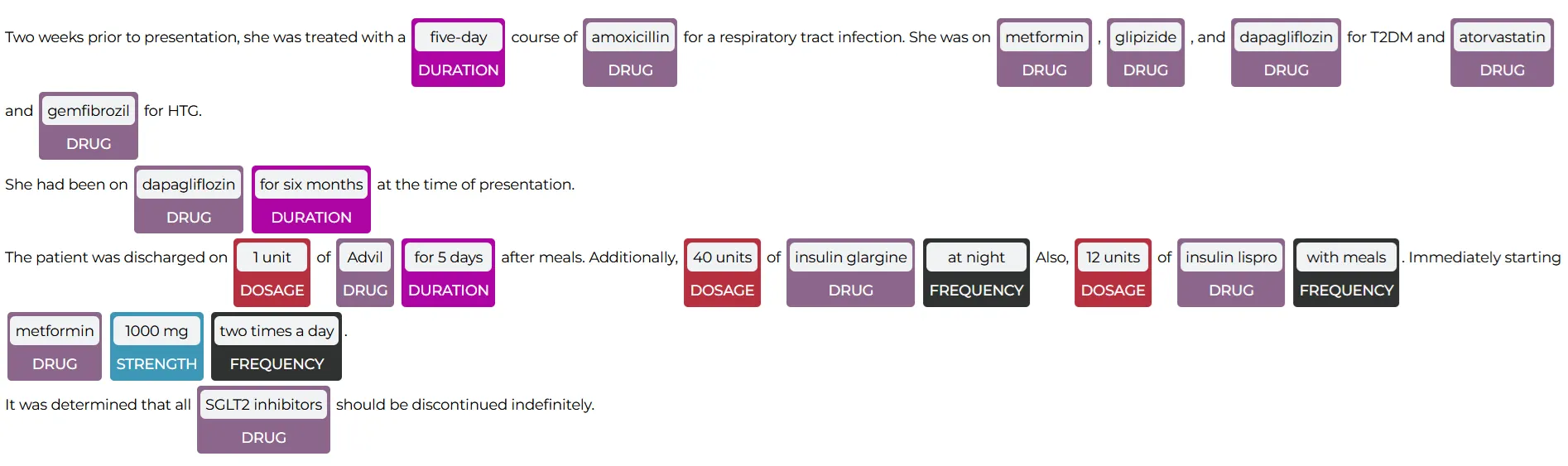
The NerVisualizer highlights the named entities that are identified by ner_posology and also displays their labels as decorations on top of the analyzed text.
Relation Extraction
The complexity of medication information often involves intricate relationships between various entities. Relation extraction models are designed to identify and map these interconnections within the text. By extracting relationships between entities, such as “DRUG” and “DOSAGE”, these models create a detailed map of the factors about medication information. This relational data is invaluable for uncovering hidden patterns and correlations that are critical for further studies.
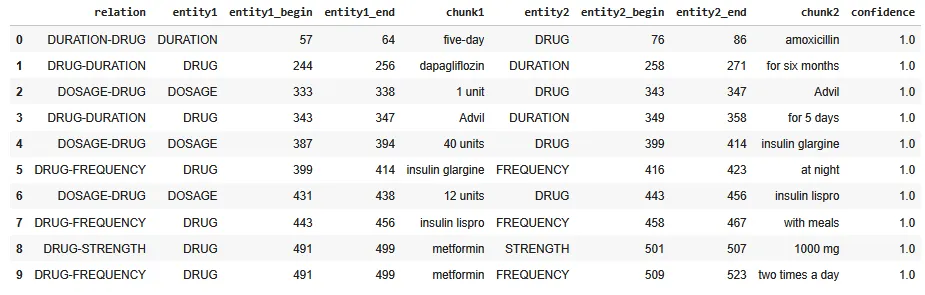
Extracted relations between entities in structured format.
John Snow Labs’ Relation Extraction Visualizer provides a powerful tool for visualizing the relationships identified by relation extraction models. This visualizer allows users to see the connections between different entities within the text and displaying these relationships in an intuitive and interactive manner helps users to better understand complex data structures, validate the accuracy of the extracted relationships, and gain deeper insights into the underlying data. This tool is particularly useful for analyzing intricate patterns and interactions in healthcare data, supporting more informed clinical and research decisions.
Knowledge Graph Generation — NetworkX
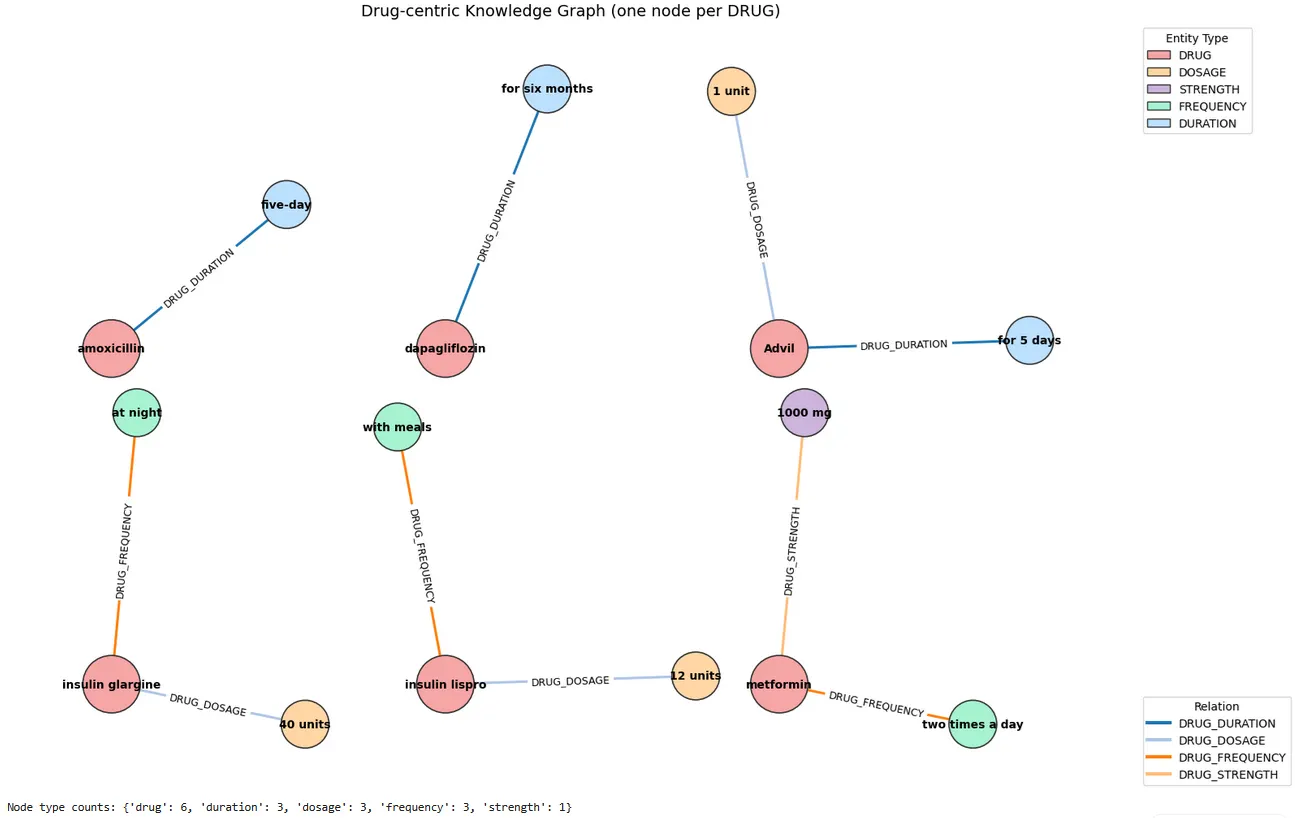
After extracting entities with the ner_posology model and linking them with the posology_re relation extractor, the final step is to materialize these outputs as a knowledge graph. At this stage, the goal is not only to store results, but to make them immediately interpretable: instead of reading a list of entities and relations, we can visually inspect how medication information is structured around each drug mention.
In the NetworkX graph, we use a drug-centric design, where each DRUG node acts as a hub and connects outward to its associated regimen attributes — DOSAGE, STRENGTH, FREQUENCY, and DURATION. This representation mirrors how clinicians think about prescriptions: a medication is meaningful only when its dose, timing, duration, and strength are attached to it. As shown in the figure, the graph separates medication “clusters” cleanly (e.g., amoxicillin → five-day, dapagliflozin → for six months, insulin glargine → 40 units → at night), making it easy to validate whether the extracted relationships are coherent at a glance.
A major advantage of this graph step is that it highlights both consistency and potential extraction errors. When the relation extractor performs well, each drug forms a compact subgraph with the expected edges (e.g., DRUG_DOSAGE, DRUG_FREQUENCY), if they are mentioned in the text. In other words, the visualization acts as a lightweight quality check before exporting the triples into a persistent graph database such as Neo4j.
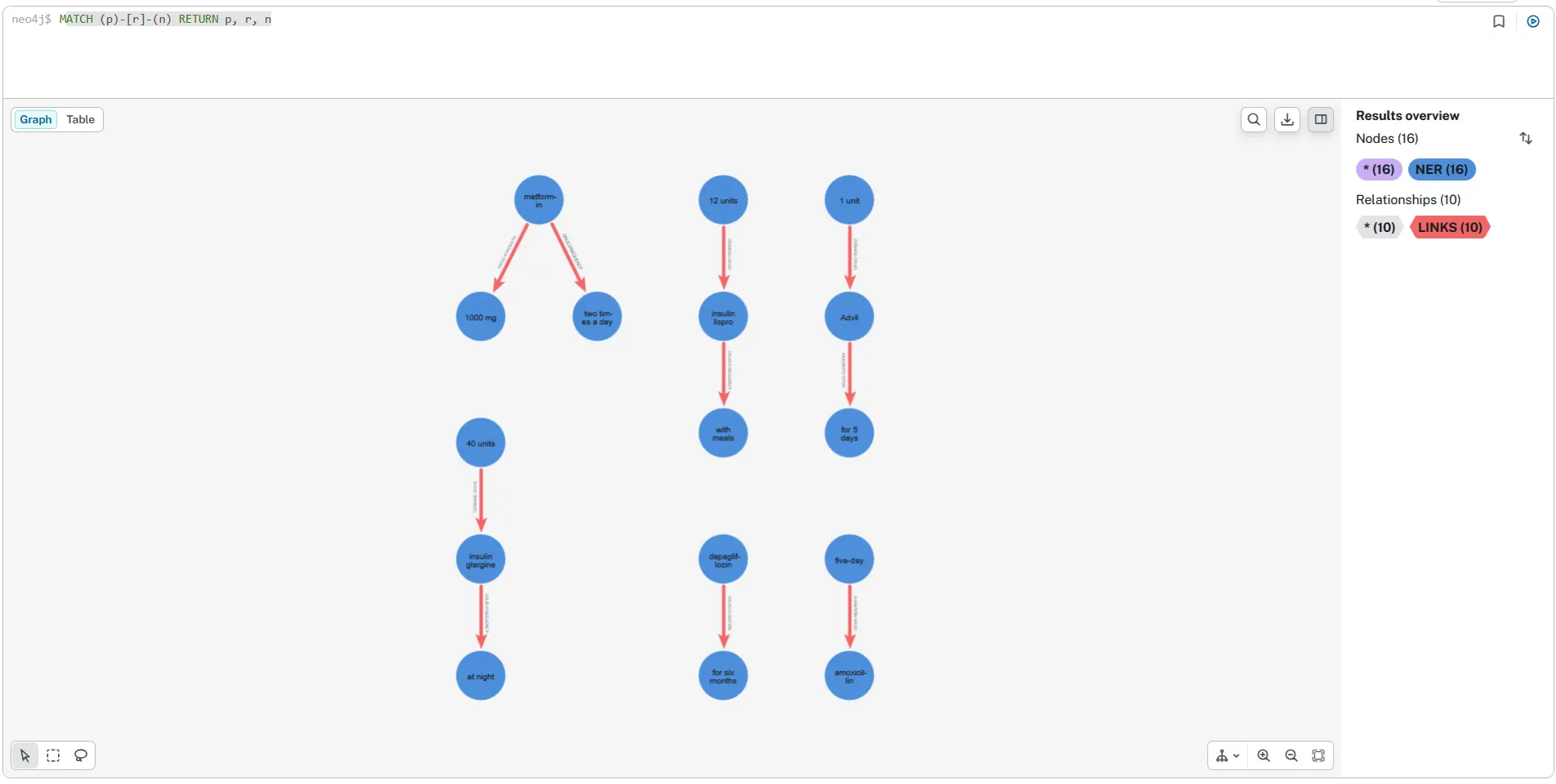
Knowledge Graph Generation — Neo4j
While NetworkX is a great way to quickly prototype and validate extracted triples in Python, a graph database like Neo4j makes the knowledge graph far more practical for exploration and downstream use. After converting the NER and relation extraction outputs into graph-friendly triples (node–edge–node), we load them into Neo4j as labeled nodes and typed relationships. This provides an interactive environment where extracted medication facts can be explored visually, filtered, and queried with Cypher. Please check the reference notebook in the Github repository for details.
The screenshots illustrate two complementary views of the same graph. The full-screen view gives a high-level overview of the extracted structure — showing multiple medication-centered clusters formed by connecting each drug to its associated attributes (dosage, frequency, duration, strength).
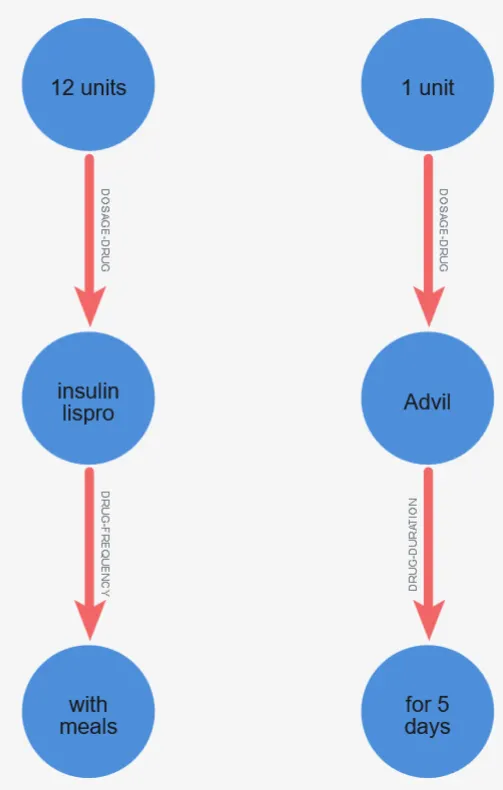
The zoomed view highlights how Neo4j helps validate the model output at a granular level: you can quickly inspect whether a specific attribute (e.g., “12 units” or “1 unit”) is attached to the correct drug and whether the relationship type matches the expected clinical meaning. This is especially useful, because graph visualization often reveals missing links or misattached attributes faster than scanning rows in a table.
Beyond visualization, Neo4j enables powerful graph queries that turn extracted NLP output into something actionable. With simple Cypher queries, you can retrieve all regimen attributes for a given drug, find drugs missing a duration or frequency, or compare dosing patterns across documents. In other words, Neo4j transforms model predictions into a persistent, queryable knowledge layer — bridging the gap between unstructured clinical narrative and structured insights that are easy to audit, search, and extend.
Conclusion
In this blog post, we showed how the John Snow Labs Healthcare NLP library can transform unstructured clinical text into structured, connected knowledge. Using a production-ready Spark NLP pipeline, we extracted key medication entities with pretrained clinical NER models, linked them with relation extraction to capture clinically meaningful connections (e.g., drug → dosage/strength/frequency/duration), and then converted those outputs into graph-ready triples. The resulting knowledge graphs — first prototyped with NetworkX and then explored interactively in Neo4j — make complex clinical narratives easier to validate, interpret, and query.
A key takeaway is that John Snow Labs Healthcare NLP is not just a collection of models — it’s a complete framework for building end-to-end clinical NLP workflows: scalable processing with Spark, high-quality pretrained medical models, configurable annotation components, and practical tooling for visualization and evaluation. At the same time, the outputs integrate naturally with the broader Python ecosystem: you can export results to Pandas for analysis, use NetworkX for rapid graph prototyping, persist graphs in Neo4j, or connect the extracted knowledge to downstream applications such as analytics pipelines, dashboards, search, and RAG-based systems. This combination — strong healthcare-native NLP plus seamless interoperability — makes it straightforward to move from clinical text to real, usable data products.